Hadoop: The Definitive Guide (2015)
Part V. Case Studies
Chapter 22. Composable Data at Cerner
Ryan Brush
Micah Whitacre
Healthcare information technology is often a story of automating existing processes. This is changing. Demands to improve care quality and control its costs are growing, creating a need for better systems to support those goals. Here we look at how Cerner is using the Hadoop ecosystem to make sense of healthcare and — building on that knowledge — to help solve such problems.
From CPUs to Semantic Integration
Cerner has long been focused on applying technology to healthcare, with much of our history emphasizing electronic medical records. However, new problems required a broader approach, which led us to look into Hadoop.
In 2009, we needed to create better search indexes of medical records. This led to processing needs not easily solved with other architectures. The search indexes required expensive processing of clinical documentation: extracting terms from the documentation and resolving their relationships with other terms. For instance, if a user typed “heart disease,” we wanted documents discussing a myocardial infarction to be returned. This processing was quite expensive — it can take several seconds of CPU time for larger documents — and we wanted to apply it to many millions of documents. In short, we needed to throw a lot of CPUs at the problem, and be cost effective in the process.
Among other options, we considered a staged event-driven architecture (SEDA) approach to ingest documents at scale. But Hadoop stood out for one important need: we wanted to reprocess the many millions of documents frequently, in a small number of hours or faster. The logic for knowledge extraction from the clinical documents was rapidly improving, and we needed to roll improvements out to the world quickly. In Hadoop, this simply meant running a new version of a MapReduce job over data already in place. The process documents were then loaded into a cluster of Apache Solr servers to support application queries.
These early successes set the stage for more involved projects. This type of system and its data can be used as an empirical basis to help control costs and improve care across entire populations. And since healthcare data is often fragmented across systems and institutions, we needed to first bring in all of that data and make sense of it.
With dozens of data sources and formats, and even standardized data models subject to interpretation, we were facing an enormous semantic integration problem. Our biggest challenge was not the size of the data — we knew Hadoop could scale to our needs — but the sheer complexity of cleaning, managing, and transforming it for our needs. We needed higher-level tools to manage that complexity.
Enter Apache Crunch
Bringing together and analyzing such disparate datasets creates a lot of demands, but a few stood out:
§ We needed to split many processing steps into modules that could easily be assembled into a sophisticated pipeline.
§ We needed to offer a higher-level programming model than raw MapReduce.
§ We needed to work with the complex structure of medical records, which have several hundred unique fields and several levels of nested substructures.
We explored a variety of options in this case, including Pig, Hive, and Cascading. Each of these worked well, and we continue to use Hive for ad hoc analysis, but they were unwieldy when applying arbitrary logic to our complex data structures. Then we heard of Crunch (seeChapter 18), a project led by Josh Wills that is similar to the FlumeJava system from Google. Crunch offers a simple Java-based programming model and static type checking of records — a perfect fit for our community of Java developers and the type of data we were working with.
Building a Complete Picture
Understanding and managing healthcare at scale requires significant amounts of clean, normalized, and relatable data. Unfortunately, such data is typically spread across a number of sources, making it difficult and error prone to consolidate. Hospitals, doctors’ offices, clinics, and pharmacies each hold portions of a person’s records in industry-standard formats such as CCDs (Continuity of Care Documents), HL7 (Health Level 7, a healthcare data interchange format), CSV files, or proprietary formats.
Our challenge is to take this data; transform it into a clean, integrated representation; and use it to create registries that help patients manage specific conditions, measure operational aspects of healthcare, and support a variety of analytics, as shown in Figure 22-1.
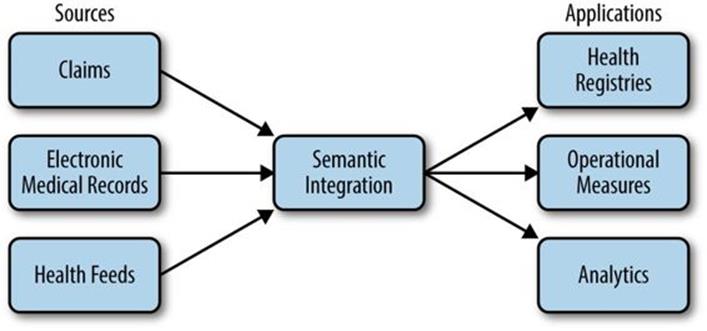
Figure 22-1. Operational data flow
An essential step is to create a clean, semantically integrated basis we can build on, which is the focus of this case study. We start by normalizing data to a common structure. Earlier versions of this system used different models, but have since migrated to Avro for storing and sharing data between processing steps. Example 22-1 shows a simplified Avro IDL to illustrate how our common structures look.
Example 22-1. Avro IDL for common data types
@namespace("com.cerner.example")
protocol PersonProtocol {
record Demographics {
string firstName;
string lastName;
string dob;
...
}
record LabResult {
string personId;
string labDate;
int labId;
int labTypeId;
int value;
}
record Medication {
string personId;
string medicationId;
string dose;
string doseUnits;
string frequency;
...
}
record Diagnosis {
string personId;
string diagnosisId;
string date;
...
}
record Allergy {
string personId;
int allergyId;
int substanceId;
...
}
/**
* Represents a person's record from a single source.
*/
record PersonRecord {
string personId;
Demographics demographics;
array<LabResult> labResults;
array<Allergy> allergies;
array<Medication> medications;
array<Diagnosis> diagnoses;
. . .
}
}
Note that a variety of data types are all nested in a common person record rather than in separate datasets. This supports the most common usage pattern for this data — looking at a complete record — without requiring downstream operations to do a number of expensive joins between datasets.
A series of Crunch pipelines are used to manipulate the data into a PCollection<PersonRecord> hiding the complexity of each source and providing a simple interface to interact with the raw, normalized record data. Behind the scenes, each PersonRecord can be stored in HDFS or as a row in HBase with the individual data elements spread throughout column families and qualifiers. The result of the aggregation looks like the data in Table 22-1.
Table 22-1. Aggregated data
|
Source |
Person ID |
Person demographics |
Data |
|
Doctor’s office |
12345 |
Abraham Lincoln ... |
Diabetes diagnosis, lab results |
|
Hospital |
98765 |
Abe Lincoln ... |
Flu diagnosis |
|
Pharmacy |
98765 |
Abe Lincoln ... |
Allergies, medications |
|
Clinic |
76543 |
A. Lincoln ... |
Lab results |
Consumers wishing to retrieve data from a collection of authorized sources call a “retriever” API that simply produces a Crunch PCollection of requested data:
Set<String> sources = ...;
PCollection<PersonRecord> personRecords =
RecordRetriever.getData(pipeline, sources);
This retriever pattern allows consumers to load datasets while being insulated from how and where they are physically stored. At the time of this writing, some use of this pattern is being replaced by the emerging Kite SDK for managing data in Hadoop. Each entry in the retrievedPCollection<PersonRecord> represents a person’s complete medical record within the context of a single source.
Integrating Healthcare Data
There are dozens of processing steps between raw data and answers to healthcare-related questions. Here we look at one: bringing together data for a single person from multiple sources.
Unfortunately, the lack of a common patient identifier in the United States, combined with noisy data such as variations in a person’s name and demographics between systems, makes it difficult to accurately unify a person’s data across sources. Information spread across multiple sources might look like Table 22-2.
Table 22-2. Data from multiple sources
|
Source |
Person ID |
First name |
Last name |
Address |
Gender |
|
Doctor’s office |
12345 |
Abraham |
Lincoln |
1600 Pennsylvania Ave. |
M |
|
Hospital |
98765 |
Abe |
Lincoln |
Washington, DC |
M |
|
Hospital |
45678 |
Mary Todd |
Lincoln |
1600 Pennsylvania Ave. |
F |
|
Clinic |
76543 |
A. |
Lincoln |
Springfield, IL |
M |
This is typically resolved in healthcare by a system called an Enterprise Master Patient Index (EMPI). An EMPI can be fed data from multiple systems and determine which records are indeed for the same person. This is achieved in a variety of ways, ranging from humans explicitly stating relationships to sophisticated algorithms that identify commonality.
In some cases, we can load EMPI information from external systems, and in others we compute it within Hadoop. The key is that we can expose this information for use in our Crunch-based pipelines. The result is a PCollection<EMPIRecord> with the data structured as follows:
@namespace("com.cerner.example")
protocol EMPIProtocol {
record PersonRecordId {
string sourceId;
string personId
}
/**
* Represents an EMPI match.
*/
record EMPIRecord {
string empiId;
array<PersonRecordId> personIds;
}
}
Given EMPI information for the data in this structure, PCollection<EMPIRecord> would contain data like that shown in Table 22-3.
Table 22-3. EMPI data
|
EMPI identifier |
PersonRecordIds (<SourceId, PersonId>) |
|
EMPI-1 |
<offc-135, 12345> |
|
EMPI-2 |
<hspt-802, 45678> |
In order to group a person’s medical records in a single location based upon the provided PCollection<EMPIRecord> and PCollection<PersonRecord>, the collections must be converted into a PTable, keyed by a common key. In this situation, a Pair<String, String>, where the first value is the sourceId and the second is the personId, will guarantee a unique key to use for joining.
The first step is to extract the common key from each EMPIRecord in the collection:
PCollection<EMPIRecord> empiRecords = ...;
PTable<Pair<String, String>, EMPIRecord> keyedEmpiRecords =
empiRecords.parallelDo(
new DoFn<EMPIRecord, Pair<Pair<String, String>, EMPIRecord>>() {
@Override
public void process(EMPIRecord input,
Emitter<Pair<Pair<String, String>, EMPIRecord>> emitter) {
for (PersonRecordId recordId: input.getPersonIds()) {
emitter.emit(Pair.of(
Pair.of(recordId.getSourceId(), recordId.getPersonId()), input));
}
}
}, tableOf(pairs(strings(), strings()), records(EMPIRecord.class)
);
Next, the same key needs to be extracted from each PersonRecord:
PCollection<PersonRecord> personRecords = ...;
PTable<Pair<String, String>, PersonRecord> keyedPersonRecords = personRecords.by(
new MapFn<PersonRecord, Pair<String, String>>() {
@Override
public Pair<String, String> map(PersonRecord input) {
return Pair.of(input.getSourceId(), input.getPersonId());
}
}, pairs(strings(), strings()));
Joining the two PTable objects will return a PTable<Pair<String, String>, Pair<EMPIRecord, PersonRecord>>. In this situation, the keys are no longer useful, so we change the table to be keyed by the EMPI identifier:
PTable<String, PersonRecord> personRecordKeyedByEMPI = keyedPersonRecords
.join(keyedEmpiRecords)
.values()
.by(new MapFn<Pair<PersonRecord, EMPIRecord>>() {
@Override
public String map(Pair<PersonRecord, EMPIRecord> input) {
return input.second().getEmpiId();
}
}, strings()));
The final step is to group the table by its key to ensure all of the data is aggregated together for processing as a complete collection:
PGroupedTable<String, PersonRecord> groupedPersonRecords =
personRecordKeyedByEMPI.groupByKey();
The PGroupedTable would contain data like that in Table 22-4.
This logic to unify data sources is the first step of a larger execution flow. Other Crunch functions downstream build on these steps to meet many client needs. In a common use case, a number of problems are solved by loading the contents of the unified PersonRecords into a rules-based processing model to emit new clinical knowledge. For instance, we may run rules over those records to determine if a diabetic is receiving recommended care, and to indicate areas that can be improved. Similar rule sets exist for a variety of needs, ranging from general wellness to managing complicated conditions. The logic can be complicated and with a lot of variance between use cases, but it is all hosted in functions composed in a Crunch pipeline.
Table 22-4. Grouped EMPI data
|
EMPI identifier |
Iterable<PersonRecord> |
|
EMPI-1 |
{ |
|
EMPI-2 |
{ |
Composability over Frameworks
The patterns described here take on a particular class of problem in healthcare centered around the person. However, this data can serve as the basis for understanding operational and systemic properties of healthcare as well, creating new demands on our ability to transform and analyze it.
Libraries like Crunch help us meet emerging demands because they help make our data and processing logic composable. Rather than a single, static framework for data processing, we can modularize functions and datasets and reuse them as new needs emerge. Figure 22-2 shows how components can be wired into one another in novel ways, with each box implemented as one or more Crunch DoFns. Here we leverage person records to identify diabetics and recommend health management programs, while using those composable pieces to integrate operational data and drive analytics of the health system.
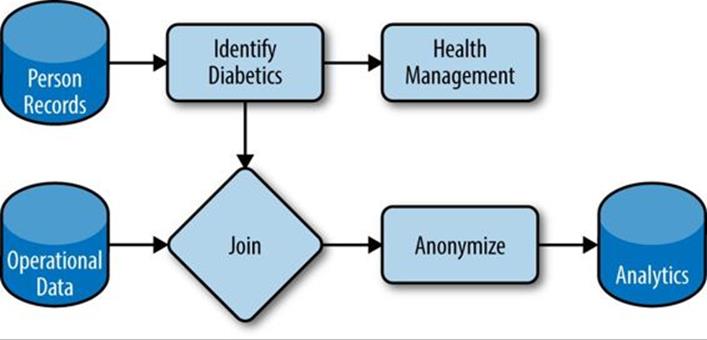
Figure 22-2. Composable datasets and functions
Composability also makes iterating through new problem spaces easier. When creating a new view of data to answer a new class of question, we can tap into existing datasets and transformations and emit our new version. As the problem becomes better understood, that view can be replaced or updated iteratively. Ultimately, these new functions and datasets can be contributed back and leveraged for new needs. The result is a growing catalog of datasets to support growing demands to understand the data.
Processing is orchestrated with Oozie. Every time new data arrives, a new dataset is created with a unique identifier in a well-defined location in HDFS. Oozie coordinators watch that location and simply launch Crunch jobs to create downstream datasets, which may subsequently be picked up by other coordinators. At the time of this writing, datasets and updates are identified by UUIDs to keep them unique. However, we are in the process of placing new data in timestamp-based partitions in order to better work with Oozie’s nominal time model.
Moving Forward
We are looking to two major steps to maximize the value from this system more efficiently.
First, we want to create prescriptive practices around the Hadoop ecosystem and its supporting libraries. A number of good practices are defined in this book and elsewhere, but they often require significant expertise to implement effectively. We are using and building libraries that make such patterns explicit and accessible to a larger audience. Crunch offers some good examples of this, with a variety of join and processing patterns built into the library.
Second, our growing catalog of datasets has created a demand for simple and prescriptive data management to complement the processing features offered by Crunch. We have been adopting the Kite SDK to meet this need in some use cases, and expect to expand its use over time.
The end goal is a secure, scalable catalog of data to support many needs in healthcare, including problems that have not yet emerged. Hadoop has shown it can scale to our data and processing needs, and higher-level libraries are now making it usable by a larger audience for manyproblems.
All materials on the site are licensed Creative Commons Attribution-Sharealike 3.0 Unported CC BY-SA 3.0 & GNU Free Documentation License (GFDL)
If you are the copyright holder of any material contained on our site and intend to remove it, please contact our site administrator for approval.
© 2016-2025 All site design rights belong to S.Y.A.