Anonymizing Health Data: Case Studies and Methods to Get You Started (2013)
Chapter 11. Masking: Oncology Databases
When we need to remove all useful data from a field, we turn to masking—the second of our pillars discussed in The Two Pillars of Anonymization. Usually this means replacing real data with entirely random values, possibly from a large database (for things like names). Obviously, this isn’t something we do to fields we need for analytics. Rather, it’s something we apply to things like names, Social Security numbers, and ID fields. De-identification involves protecting fields we need for analytics, and is a trade-off between privacy and utility; masking involves protecting fields we don’t need for analytics, and is meant to completely hide the original data.
To understand the reasons for masking and its trade-offs, we’ll take a short look at a real database. The American Society of Clinical Oncology (ASCO) has launched an ambitious project to build tools on top of oncology electronic health record (EHR) data collected from sites across the country. Its goal is to improve the quality of care by having millions of patients essentially participate in a large clinical trial, pooling all of their data in a system called CancerLinQ.[80]
Schema Shmema
Before we discuss data masking, let’s look at an example database that the ASCO CancerLinQ system may come across. This will give us examples to think about when we go through approaches to masking. Figure 11-1 is a schema for our invented database. Direct identifiers include the names, address (although we’ll keep the ZIP code as a quasi-identifier), and SSN. But the patient ID is unique to the patient, at least within the EHR, so it also has to be masked (better safe than sorry). Short free-form text fields might contain identifying information, like the patient’s name, so unless we’re going to de-identify that we’ll need to mask it as well.
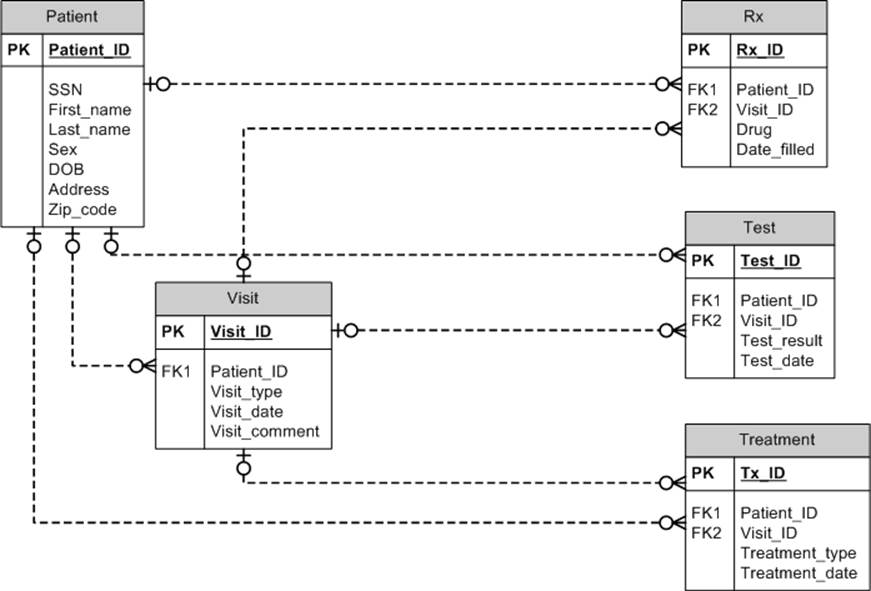
Figure 11-1. Like a school of herring, these tables stick together. Of course, this is only one example schema for an oncology database; there could be many variations.
Data in Disguise
There are three defensible approaches to data masking. If implemented properly, they will ensure that the risk of re-identification is extremely small. And because these approaches will, by definition, ensure small risk, it’s not necessary to evaluate risk levels for masking. Just implementing the recommended masking schemes is sufficient.
Field Suppression
Suppression is by far the simplest way to mask data—just remove the direct identifiers from the data set. This could be done with the first name, last name, and SSN, to name a few. It’s perfect for those pesky comment fields we mentioned in the oncology database, since we don’t know what’s in them and really, since they’re just short notes, we don’t need them. If we did, we would turn to the methods to anonymize free-form text that we discussed in Chapter 8.
Suppression is an acceptable approach in some situations. If we’re disclosing a data set to a researcher, it’s pretty unlikely that he’ll need any of these fields. An exception would be if the researcher were planning to contact the patients; then having their names would help. But then it wouldn’t be an anonymized data set.
Another situation that often comes up is when the researcher wants to link the data set with another one, so the SSN is needed for the purpose of linking. But we don’t want to disclose SSN values in the clear because it’s personal information. We’ll see how to get around this in Chapter 12, where we describe a method for linking on unique identifiers, such as the SSN, without revealing the actual values to the researcher.
But suppression isn’t always acceptable. When developing health applications, there’s a need to test software. It’s necessary to work with realistic data to perform meaningful functional and performance testing of applications. The most realistic data is real data, coming from live production systems. But you do not want to give your development teams real patient data.
Software testing would definitely be considered a secondary use of the data. This raises all of the usual regulatory issues around using identifiable data for secondary purposes without consent. But there are also known, and well-publicized, cases of the development team losing patient data.[81]
When testing a software application, it’s necessary to maintain all of the fields in that database in their original format. You can’t suppress entire fields because the data must fill in all of the database columns to facilitate testing. Field suppression is simply not an option here.
A related example is when users of health applications run into problems with the software, possibly due to bugs. They’ll want to send you some sample data to recreate the problem, so that your development team can fix it quickly. You would want to mask any identifying data before sending it to the development team, but you wouldn’t want to suppress identifying fields in case they’re causing the problem. Similar examples include use for software demos and training.
Randomization
Randomization is the process of replacing the direct identifier values with fake data: fake person names, for example, or fake SSNs. The fake values can be generated independently of the data, using random characters for names and random numbers for SSNs. Or the fake values can be selected randomly from a large database of known values, such as a database of real first names and last names. Randomization can result in some very real-looking data sets.
NOTE
If values are being selected from a database, the database should be large enough to render the probability of selecting any one name very small. Randomization can also be performed to enforce uniqueness: for example, replace every name with a unique name. We definitely need to enforce uniqueness when randomizing patient and facility IDs in the oncology database; otherwise, relationships in the data will be lost.
Some conditions can be placed on the randomization. A perfect example is tying names to the patient’s sex, so that patients are assigned more or less sex-specific names. It would make sense to use typically female names for breast cancer data, or typically male names for prostate cancer data. But you have to be careful about how specific the conditions should be. You don’t want to give away information, so the conditions that you match to should be in the de-identified data. If you’ve dropped the patient’s sex from the data set, forget about matching name to sex. Otherwise, the name would leak information about the patient’s sex.
Replacing names with random names that have the same length would also be a bad idea (due to the possibility of a frequency attack). Some surnames have 20 characters, but only a handful, and maybe only in a particular geography. If an adversary can know the length of the name, she might be able to figure out the original name. And it’s extremely unlikely that the length of a person’s name is otherwise included in the data set. (If so, then include it as a quasi-identifier, although we’ve never heard of such a thing!)
We looked at the North Carolina voter registration list, which is available publicly, and found that there were only two people with first names having 17 characters, four people with last names having 16 characters, and three people with last names having 21 characters. We also looked at a registry of lawyers and doctors in Ontario, and there was only person with a first name having 17, 18, or 20 characters, and only one person with a last name having 24 characters.
It’s possible to include ethnicity in a de-identified data set, so this could be used as a condition on which names are randomized. For example, if there’s a field indicating that a patient is Asian, then an Asian name would be used in the randomization. But otherwise, it’s a bad idea to use this strategy to randomize names, as it will inadvertently increase the risk of re-identification.
WARNING
When you use both masking and de-identification on a data set, any masking that depends on a quasi-identifier needs to be applied after de-identification. So, if the first name is being randomized on the condition of the patient’s sex, make sure you do the masking on the de-identified sex field so that suppression is taken into account. Otherwise, you might leak the sex of the patient where you had intended to have it removed!
When randomizing fields in a relational database, ensuring consistency is important. The schema in Figure 11-1 is normalized, so the names do not appear in multiple tables. But there are many databases out there that are not normalized—e.g., the First_name and Last_name fields might appear in other tables as well. If the Last_name is randomized to “Herring” from “Salmon,” then all instances of Last_name for that patient in other tables will also be converted to “Herring.”
Pseudonymization
Pseudonymization replaces one or more direct identifiers with some other unique value. For example, an SSN may be replaced with another fake SSN value. We consider pseudonyms to be derived from the original value. Therefore, if we have an SSN and we generate a pseudonym from it repeatedly, we will always get the same pseudonym value. This is an important characteristic of pseudonyms because it allows the matching of patient records across time.
Suppose we have a database of patients that we’re using for testing and we create pseudonyms from their SSNs. Then, after six months, we get an update to that database. Because the pseudonym generated for each patient will be the same in the original database and the new increment, it will be possible to determine which records belong to the same patient.
Two general methods are used to generate pseudonyms: hashing and encryption. With hashing it’s necessary to add a random value called a salt to the SSN to foil dictionary attacks on the pseudonym. This salt must not be shared with any outsider, because it’s essentially a key to unlock the pseudonym.
The disadvantage of hashing is that it results in a large value: 32, 44, or 64 bytes when converted to an ASCII string. If our database has allocated 11 characters for an SSN field, we won’t be able to stuff the hash of that SSN into the field. Recall also that in some situations, such as software testing, we want to maintain the integrity of the database, which includes all of the fields and their types.
Format-preserving encryption is a type of encryption that will create an encrypted value that can fit into the same field length as the original value. It also uses a secret key that must be kept in a safe place. It solves the problem of ensuring that the database schema is not distorted after the pseudonym is created.
Referential integrity is also an important capability for pseudonyms, because these pseudonyms are often used to link individuals’ records across tables. This can be maintained in the same manner as for randomization schemes. If an identifier is converted to a pseudonym in one table, then it will need to be converted in the same manner in all tables in the database. In our schema in Figure 11-1, the Patient_ID might be converted to a pseudonym in the Patient table. But to ensure that the tables can be joined properly, that same Patient_ID would need to be converted to the same pseudonym in other tables, such as the Visit table or the Rx table.
CONTROLLED RE-IDENTIFICATION
In some cases it’s necessary to determine the identity of the data subject. For example, in a public health context it may be determined that an individual has been exposed to a substance or pathogen that requires follow-up, or that contact tracing is required. In those cases it may be necessary to reverse the pseudonyms created.
WARNING
Reversing pseudonyms needs to be performed in a secure environment, with appropriate controls and oversight, and under conditions that have been defined in advance (not willy-nilly, when the mood arises). You need to be sure that the re-identification of individuals is legitimate.
To allow the reversing of pseudonyms that are hash values, you need to maintain a mapping table between the pseudonyms and the original values. This mapping table needs to be kept in a secure place and referred to only when a re-identification is necessary.
When you have used format-preserving encryption, the only thing you need to store securely is the encryption key. The key can then be used to decrypt the pseudonym and recover the original values. The advantage here is that a whole mapping table doesn’t need to be kept. For large data sets, this mapping table can grow significantly in size.
Frequency of Pseudonyms
We have to be careful with pseudonyms, as they could still leak information through their frequency. If unique identifiers, such as MRNs or SSNs, are converted to pseudonyms using the methods we describe, then there shouldn’t be leakage of information. However, sometimes pseudonyms are created for other types of fields, such as facility names or even regions.
Consider a data set where there’s a field indicating the hospital where care was provided. As part of data masking the hospital name was converted to a pseudonym. This conversion seems to hide the hospital name, but allows an analyst to know which patients went to the same hospital. So far, so good.
Now, in the city we live in, Ottawa, there’s one very large hospital and a few small hospitals. Most adult patients go The Ottawa Hospital (TOH). Any data set of patients discharged from Ottawa hospitals will have a high frequency for the pseudonym of TOH. It doesn’t matter what the pseudonym is; its frequency will make it obvious which pseudonym is for TOH. Similarly, there’s only one children’s hospital in the region, and the hospital pseudonym that has the highest frequency of pediatric discharges will most likely be for the children’s hospital.
An adversary can therefore reverse engineer the hospital using only frequency information. Performing a test to determine if the pseudonym distribution deviates too much from a uniform distribution should reveal these types of situations.
A better approach is to dichotomize that field into “TOH Patients” and “Non-TOH Patients,” then include that new dichotomous field in the usual re-identification risk assessment (effectively treating it as a quasi-identifier). If a particular value is suppressed, then that pseudonym is also suppressed. If a value in the new dichotomous field is not suppressed, then it’s OK to reveal that pseudonym. The nesting can be more complex, though.
This isn’t just for hospitals—there are specialty centers in Ottawa as well (there’s a heart institute, a cancer center, and an eye institute). So, these concerns apply to the frequencies within a particular diagnosis group as well.
Masking On the Fly
Masking functionality can be embedded between the database and the presentation layer of an application. The presentation layer may be a summary table from a data set, or a data export. When the data from the database is pulled out to be processed, the fields that are designated as direct identifiers are then masked automatically and the masked values are transmitted on to the next layer in the application.
On-the-fly masking is being considered for the CancerLinQ system that ASCO is developing, depending on user access privileges. It’s currently still in the prototype phase, so this is not implemented, but it would certainly help give different users different layers of access.
Generally, this kind of approach works well in environments where the data is dynamic, changing on a regular basis. It’s also suitable when the database scheme is fixed and the direct identifiers can be determined in advance. Data users are assured that any data extracted from the database is always masked. The automated masking eliminates the need for manual interventions to ensure that the data is properly masked.
Of course, the setup for on-the-fly masking would need to be updated every time the database scheme was changed (for example, if a new direct identifier was added to the database).
Final Thoughts
It should be clear in reading this book that masking alone is not nearly enough to protect the identity of an individual. Masking is necessary, but it’s not sufficient. The other methods that we’ve presented for de-identification must also be used. The reason we reiterate this point is that we still see organizations that are handling sensitive health information who do only masking without de-identification. These organizations are taking a nontrivial risk, but if you’re reading this book, you know better.
[80] R. Winslow, " ‘Big Data’ for Cancer Care: Vast Storehouse of Patient Records Will Let Doctors Search for Effective Treatment,” The Wall Street Journal, 27 March 2013.
[81] S. Rubenstein, “Express Scripts Data Breach Leads to Extortion Attempt,” The Wall Street Journal, 7 November 2008.