Functional Python Programming (2015)
Chapter 8. The Itertools Module
Functional programming emphasizes stateless programming. In Python this leads us to work with generator expressions, generator functions, and iterables. In this chapter, we'll look at the itertools library with numerous functions to help us work with iterable collections.
We introduced iterator functions in Chapter 3, Functions, Iterators, and Generators. In this chapter, we'll expand on that superficial introduction. We used some related functions in Chapter 5, Higher-order Functions.
Note
Some of the functions merely behave like they are proper, lazy Python iterables. It's important to look at the implementation details for each of these functions. Some of them create intermediate objects, leading to the potential of consuming a large amount of memory. Since implementations might change with Python releases, we can't provide function-by-function advice here. If you have performance or memory issues, ensure that you check the implementation.
There are a large number of iterator functions in this module. We'll examine some of the functions in the next chapter. In this chapter, we'll look at three broad groupings of iterator functions. These are as follows:
· Functions that work with infinite iterators. These can be applied to any iterable or an iterator over any collection; they will consume the entire source.
· Functions that work with finite iterators. These can either accumulate a source multiple times, or they produce a reduction of the source.
· The tee iterator function which clones an iterator into several copies that can each be used independently. This provides a way to overcome the primary limitation of Python iterators: they can be used once only.
We need to emphasize an important limitation of iterables that we've touched upon in other places.
Note
Iterables can be used only once.
This can be astonishing because there's no error. Once exhausted, they appear to have no elements and will raise the StopIteration exception every time they're used.
There are some other features of iterators that aren't such profound limitations. They are as follows:
· There's no len() function for an iterable. In almost every other respect, they appear to be a container.
· Iterables can do next() operations, unlike a container.
· The for statement makes the distinction between containers and iterables invisible; containers will produce an iterable via the iter() function. An iterable simply returns itself.
These points will provide some necessary background for this chapter. The idea of the itertools module is to leverage what iterables can do to create succinct, expressive applications without the complex-looking overheads associated with the details of managing the iterables.
Working with the infinite iterators
The itertools module provides a number of functions that we can use to enhance or enrich an iterable source of data. We'll look at the following three functions:
· count(): This is an unlimited version of the range() function
· cycle(): This will reiterate a cycle of values
· repeat(): This can repeat a single value an indefinite number of times
Our goal is to understand how these various iterator functions can be used in generator expressions and with generator functions.
Counting with count()
The built-in range() function is defined by an upper limit: the lower limit and step values are optional. The count() function, on the other hand, has a start and optional step, but no upper limit.
This function can be thought of as the primitive basis for a function like enumerate(). We can define the enumerate() function in terms of zip() and count() functions, as follows:
enumerate = lambda x, start=0: zip(count(start),x)
The enumerate() function behaves as if it's a zip() function that uses the count() function to generate the values associated with some iterator.
Consequently, the following two commands are equivalent to each other:
zip(count(), some_iterator)
enumerate(some_iterator)
Both will emit a sequence of numbers of two tuples paired with items from the iterator.
The zip() function is made slightly simpler with the use of the count() function, as shown in the following command:
zip(count(1,3), some_iterator)
This will provide values of 1, 4, 7, 10, and so on, as the identifiers for each value from the enumerator. This is a challenge because enumerate doesn't provide a way to change the step.
The following command describes the enumerate() function:
((1+3*e, x) for e,x in enumerate(a))
Note
The count() function permits non-integer values. We can use something like the count(0.5, 0.1) method to provide floating-point values. This will accumulate a substantial error if the increment value doesn't have an exact representation. It's generally better to use the (0.5+x*.1 for x in count()) method to assure that representation errors don't accumulate.
Here's a way to examine the accumulating error. We'll define a function, which will evaluate items from an iterator until some condition is met. Here's how we can define the until() function:
def until(terminate, iterator):
i = next(iterator)
if terminate(*i): return i
return until(terminate, iterator)
We'll get the next value from the iterator. If it passes the test, that's our value. Otherwise, we'll evaluate this function recursively to search for a value that passes the test.
We'll provide a source iterable and a comparison function as follows:
source = zip(count(0, .1), (.1*c for c in count()))
neq = lambda x, y: abs(x-y) > 1.0E-12
When we evaluate the until(neq, source) method, we find the result is as follows:
(92.799999999999, 92.80000000000001)
After 928 iterations, the sum of the error bits has accumulated to ![]() . Neither value has an exact binary representation.
. Neither value has an exact binary representation.
Note
The count() function is close to the Python recursion limit. We'd need to rewrite our until() function to use tail-call optimization to locate counts with larger accumulated errors.
The smallest detectible difference can be computed as follows:
>>> until(lambda x, y: x != y, source)
(0.6, 0.6000000000000001)
After just six steps, the count(0, 0.1) method has accumulated a measurable error of ![]() . Not a large error, but within 1000 steps, it will be considerably larger.
. Not a large error, but within 1000 steps, it will be considerably larger.
Reiterating a cycle with cycle()
The cycle() function repeats a sequence of values. We can imagine using it to solve silly fizz-buzz problems.
Visit http://rosettacode.org/wiki/FizzBuzz for a comprehensive set of solutions to a fairly trivial programming problem. Also see https://projecteuler.net/problem=1 for an interesting variation on this theme.
We can use the cycle() function to emit sequences of True and False values as follows:
m3= (i == 0 for i in cycle(range(3)))
m5= (i == 0 for i in cycle(range(5)))
If we zip together a finite collection of numbers, we'll get a set of triples with a number, and two flags showing whether or not the number is a multiple of 3 or a multiple of 5. It's important to introduce a finite iterable to create a proper upper bound on the volume of data being generated. Here's a sequence of values and their multiplier flags:
multipliers = zip(range(10), m3, m5)
We can now decompose the triples and use a filter to pass numbers which are multiples and reject all others:
sum(i for i, *multipliers in multipliers if any(multipliers))
This function has another, more valuable use for exploratory data analysis.
We often need to work with samples of large sets of data. The initial phases of cleansing and model creation are best developed with small sets of data and tested with larger and larger sets of data. We can use the cycle() function to fairly select rows from within a larger set. The population size, ![]() , and the desired sample size,
, and the desired sample size, ![]() , denotes how long we can use a cycle:
, denotes how long we can use a cycle:
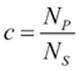
We'll assume that the data can be parsed with the csv module. This leads to an elegant way to create subsets. We can create subsets using the following commands:
chooser = (x == 0 for x in cycle(range(c)))
rdr= csv.reader(source_file)
wtr= csv.writer(target_file)
wtr.writerows(row for pick, row in zip(chooser, rdr) if pick)
We created a cycle() function based on the selection factor, c. For example, we might have a population of 10 million records: a 1,000-record subset involves picking 1/10,000 of the records. We assumed that this snippet of code is nestled securely inside a withstatement that opens the relevant files. We also avoided showing details of any dialect issues with the CSV format files.
We can use a simple generator expression to filter the data using the cycle() function and the source data that's available from the CSV reader. Since the chooser expression and the expression used to write the rows are both non-strict, there's little memory overhead from this kind of processing.
We can—with a small change—use the random.randrange(c) method instead of the cycle(c) method to achieve a randomized selection of a similar sized subset.
We can also rewrite this method to use compress(), filter(), and islice() functions, as we'll see later in this chapter.
This design will also reformat a file from any nonstandard CSV-like format into a standardized CSV format. As long as we define parser functions that return consistently defined tuples and write consumer functions that write tuples to the target files, we can do a great deal of cleansing and filtering with relatively short, clear scripts.
Repeating a single value with repeat()
The repeat() function seems like an odd feature: it returns a single value over and over again. It can serve as a replacement for the cycle() function. We can extend our data subset selection function using the repeat(0) method instead of the cycle(range(100))method in an expression line, for example,(x==0 for x in some_function).
We can think of the following commands:
all = repeat(0)
subset= cycle(range(100))
chooser = (x == 0 for x in either_all_or_subset)
This allows us to make a simple parameter change, which will either pick all data or pick a subset of data.
We can embed this in nested loops to create more complex structures. Here's a simple example:
>>> list(tuple(repeat(i, times=i)) for i in range(10))
[(), (1,), (2, 2), (3, 3, 3), (4, 4, 4, 4), (5, 5, 5, 5, 5), (6, 6, 6, 6, 6, 6), (7, 7, 7, 7, 7, 7, 7), (8, 8, 8, 8, 8, 8, 8, 8), (9, 9, 9, 9, 9, 9, 9, 9, 9)]
>>> list(sum(repeat(i, times=i)) for i in range(10))
[0, 1, 4, 9, 16, 25, 36, 49, 64, 81]
We created repeating sequences of numbers using the times parameter on the repeat() function.
Using the finite iterators
The itertools module provides a number of functions that we can use to produce finite sequences of values. We'll look at ten functions in this module, plus some related built-in functions:
· enumerate(): This function is actually part of the __builtins__ package, but it works with an iterator and is very similar to other functions in the itertools module.
· accumulate(): This function returns a sequence of reductions of the input iterable. It's a higher-order function and can do a variety of clever calculations.
· chain(): This function combines multiple iterables serially.
· groupby(): This function uses a function to decompose a single iterable into a sequence of iterables over subsets of the input data.
· zip_longest(): This function combines elements from multiple iterables. The built-in zip() function truncates the sequence at the length of the shortest iterable. The zip_longest() function pads the shorter iterables with the given fillvalue.
· compress(): This function filters one iterable based on a second iterable of Boolean values.
· islice(): This function is the equivalent of a slice of a sequence when applied to an iterable.
· dropwhile() and takewhile(): Both of these functions use a Boolean function to filter items from an iterable. Unlike filter() or filterfalse(), these functions rely on a single True or False value to change their filter behavior for all subsequent values.
· filterfalse(): This function applies a filter function to an iterable. This complements the built-in filter() function.
· starmap(): This function maps a function to an iterable sequence of tuples using each iterable as an *args argument to the given function. The map() function does a similar thing using multiple parallel iterables.
We've grouped these functions into approximate categories. The categories are roughly related to concepts of restructuring an iterable, filtering, and mapping.
Assigning numbers with enumerate()
In Chapter 7, Additional Tuple Techniques, we used the enumerate() function to make a naïve assignment of rank numbers to sorted data. We can do things like pairing up a value with its position in the original sequence, as follows:
pairs = tuple(enumerate(sorted(raw_values)))
This will sort the items in raw_values into order, create two tuples with an ascending sequence of numbers, and materialize an object we can use for further calculations. The command and the result are as follows:
>>> raw_values= [1.2, .8, 1.2, 2.3, 11, 18]
>>> tuple(enumerate( sorted(raw_values)))
((0, 0.8), (1, 1.2), (2, 1.2), (3, 2.3), (4, 11), (5, 18))
In Chapter 7, Additional Tuple Techniques we implemented an alternative form of enumerate, rank() function, which would handle ties in a more statistically useful way.
This is a common feature that is added to a parser to record the source data row numbers. In many cases, we'll create some kind of row_iter() function to extract the string values from a source file. This might iterate over the string values in tags of an XML file or in columns of a CSV file. In some cases, we might even be parsing data presented in an HTML file parsed with Beautiful Soup.
In Chapter 4, Working with Collections, we parsed an XML to create a simple sequence of position tuples. We then created legs with a start, end, and distance. We did not, however, assign an explicit leg number. If we ever sorted the trip collection, we'd be unable to determine the original ordering of the legs.
In Chapter 7, Additional Tuple Techniques, we expanded on the basic parser to create namedtuples for each leg of the trip. The output from this enhanced parser looks as follows:
(Leg(start=Point(latitude=37.54901619777347, longitude=-76.33029518659048), end=Point(latitude=37.840832, longitude=-76.273834), distance=17.7246), Leg(start=Point(latitude=37.840832, longitude=-76.273834), end=Point(latitude=38.331501, longitude=-76.459503), distance=30.7382), Leg(start=Point(latitude=38.331501, longitude=-76.459503), end=Point(latitude=38.845501, longitude=-76.537331), distance=31.0756),...,Leg(start=Point(latitude=38.330166, longitude=-76.458504), end=Point(latitude=38.976334, longitude=-76.473503), distance=38.8019))
The first Leg function is a short trip between two points on the Chesapeake Bay.
We can add a function that will build a more complex tuple with the input order information as part of the tuple. First, we'll define a slightly more complex version of the Leg class:
Leg = namedtuple("Leg", ("order", "start", "end", "distance"))
This is similar to the Leg instance shown in Chapter 7, Additional Tuple Techniques but it includes the order as well as the other attributes. We'll define a function that decomposes pairs and creates Leg instances as follows:
def ordered_leg_iter(pair_iter):
for order, pair in enumerate(pair_iter):
start, end = pair
yield Leg(order, start, end, round(haversine(start, end),4))
We can use this function to enumerate each pair of starting and ending points. We'll decompose the pair and then reassemble the order, start, and end parameters and the haversine(start,end) parameter's value as a single Leg instance. This generator function will work with an iterable sequence of pairs.
In the context of the preceding explanation, it is used as follows:
with urllib.request.urlopen("file:./Winter%202012-2013.kml") as source:
path_iter = float_lat_lon(row_iter_kml(source))
pair_iter = legs(path_iter)
trip_iter = ordered_leg_iter(pair_iter)
trip= tuple(trip_iter)
We've parsed the original file into the path points, created start-end pairs, and then created a trip that was built of individual Leg objects. The enumerate() function assures that each item in the iterable sequence is given a unique number that increments from the default starting value of 0. A second argument value can be given to provide an alternate starting value.
Running totals with accumulate()
The accumulate() function folds a given function into an iterable, accumulating a series of reductions. This will iterate over the running totals from another iterator; the default function is operator.add(). We can provide alternative functions to change the essential behavior from sum to product. The Python library documentation shows a particularly clever use of the max() function to create a sequence of maximum values so far.
One application of running totals is quartiling data. We can compute the running total for each sample and divide them into quarters with an int(4*value/total) calculation.
In the Assigning numbers with enumerate() section, we introduced a sequence of latitude-longitude coordinates that describe a sequence of legs on a voyage. We can use the distances as a basis for quartiling the waypoints. This allows us to determine the midpoint in the trip.
The value of the trip variable looks as follows:
(Leg(start=Point(latitude=37.54901619777347, longitude=-76.33029518659048), end=Point(latitude=37.840832, longitude=-76.273834), distance=17.7246), Leg(start=Point(latitude=37.840832, longitude=-76.273834), end=Point(latitude=38.331501, longitude=-76.459503), distance=30.7382), ..., Leg(start=Point(latitude=38.330166, longitude=-76.458504), end=Point(latitude=38.976334, longitude=-76.473503), distance=38.8019))
Each Leg object has a start point, an end point, and a distance. The calculation of quartiles looks like the following example:
distances= (leg.distance for leg in trip)
distance_accum= tuple(accumulate(distances))
total= distance_accum[-1]+1.0
quartiles= tuple(int(4*d/total) for d in distance_accum)
We extracted the distance values and computed the accumulated distances for each leg. The last of the accumulated distances is the total. We've added 1.0 to the total to assure that 4*d/total is 3.9983, which truncates to 3. Without the +1.0, the final item would have a value of 4, which is an impossible fifth quartile. For some kinds of data (with extremely large values) we might have to add a larger value.
The value of the quartiles variable is as follows:
(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3)
We can use the zip() function to merge this sequence of quartile numbers with the original data points. We can also use functions like groupby() to create distinct collections of the legs in each quartile.
Combining iterators with chain()
We can use the chain() function to combine a collection of iterators into a single, overall iterator. This can be helpful to combine data that was decomposed via the groupby() function. We can use this to process a number of collections as if they were a single collection.
In particular, we can combine the chain() function with the contextlib.ExitStack() method to process a collection of files as a single iterable sequence of values. We can do something like this:
from contextlib import ExitStack
import csv
def row_iter_csv_tab(*filenames):
with ExitStack() as stack:
files = [stack.enter_context(open(name, 'r', newline=''))
for name in filenames]
readers = [csv.reader(f, delimiter='\t') for f in files]
readers = map(lambda f: csv.reader(f, delimiter='\t'), files)
yield from chain(*readers)
We've created an ExitStack object that can contain a number of individual contexts open. When the with statement finishes, all items in the ExitStack object will be closed properly. We created a simple sequence of open file objects; these objects were also entered into the ExitStack object.
Given the sequence of files in the files variable, we created a sequence of CSV readers in the readers variable. In this case, all of our files have a common tab-delimited format, which makes it very pleasant to open all of the files with a simple, consistent application of a function to the sequence of files.
We could also open the files using the following command:
readers = map(lambda f: csv.reader(f, delimiter='\t'), files)
Finally, we chained all of the readers into a single iterator with chain(*readers). This was used to yield the sequence of rows from all of the files.
It's important to note that we can't return the chain(*readers) object. If we do, this would exit the with statement context, closing all the source files. Instead, we must yield individual rows so that the with statement context is kept active.
Partitioning an iterator with groupby()
We can use the groupby() function to partition an iterator into smaller iterators. This works by evaluating the given key() function for each item in the given iterable. If the key value matches the previous item's key, the two items are part of the same partition. If the key does not match the previous item's key, the previous partition is ended and a new partition is started.
The output from the groupby() function is a sequence of two tuples. Each tuple has the group's key value and an iterable over the items in the group. Each group's iterator can be preserved as a tuple or processed to reduce it to some summary value. Because of the way the group iterators are created, they can't be preserved.
In the Running totals with accumulate() section, earlier in the chapter, we showed how to compute quartile values for an input sequence.
Given the trip variable with the raw data and the quartile variable with the quartile assignments, we can group the data using the following commands:
group_iter= groupby(zip(quartile, trip), key=lambda q_raw:
q_raw[0])
for group_key, group_iter in group_iter:
print(group_key, tuple(group_iter))
This will start by zipping the quartile numbers with the raw trip data, iterating over two tuples. The groupby() function will use the given lambda variable to group by the quartile number. We used a for loop to examine the results of the groupby() function. This shows how we get a group key value and an iterator over members of the group.
The input to the groupby() function must be sorted by the key values. This will assure that all of the items in a group will be adjacent.
Note that we can also create groups using the defaultdict(list) method, as follows:
def groupby_2(iterable, key):
groups = defaultdict(list)
for item in iterable:
groups[key(item)].append(item)
for g in groups:
yield iter(groups[g])
We created a defaultdict class with a list object as the value associated with each key. Each item will have the given key() function applied to create a key value. The item is appended to the list in the defaultdict class with the given key.
Once all of the items are partitioned, we can then return each partition as an iterator over the items that share a common key. This is similar to the groupby() function because the input iterator to this function isn't necessarily sorted in precisely the same order; it's possible that the groups might have the same members, but the order might differ.
Merging iterables with zip_longest() and zip()
We saw the zip() function in Chapter 4, Working with Collections. The zip_longest() function differs from the zip() function in an important way: where the zip() function stops at the end of the shortest iterable, the zip_longest() function pads short iterables and stops at the end of the longest iterable.
The fillvalue keyword parameter allows filling with a value other than the default value, None.
For most exploratory data analysis applications, padding with a default value is statistically difficult to justify. The Python Standard Library document shows a few clever things which can be done with the zip_longest() function. It's difficult to expand on these without drifting far from our focus on data analysis.
Filtering with compress()
The built-in filter() function uses a predicate to determine if an item is passed or rejected. Instead of a function that calculates a value, we can use a second, parallel iterable to determine which items to pass and which to reject.
We can think of the filter() function as having the following definition:
def filter(iterable, function):
i1, i2 = tee(iterable, 2)
return compress(i1, (function(x) for x in i2))
We cloned the iterable using the tee() function. (We'll look at this function in detail later.) We evaluated the filter predicate for each value. Then we provided the original iterable and the filter function iterable to compress, pass, and reject values. This builds the features of the filter() function from the more primitive features of the compress() function.
In the Reiterating a cycle with cycle() section of this chapter, we looked at data selection using a simple generator expression. Its essence was as follows:
chooser = (x == 0 for x in cycle(range(c)))
keep= (row for pick, row in zip(chooser, some_source) if pick)
We defined a function which would produce a value 1 followed by c-1 zeroes. This cycle would be repeated, allowing to pick only 1/c rows from the source.
We can replace the cycle(range(c)) function with the repeat(0) function to select all rows. We can also replace it with the random.randrange(c) function to randomize the selection of rows.
The keep expression is really just a compress(some_source, chooser) method. If we make that change, the processing is simplified:
all = repeat(0)
subset = cycle(range(c))
randomized = random.randrange(c)
selection_rule = one of all, subset, or randomized
chooser = (x == 0 for x in selection_rule)
keep = compress(some_source, chooser)
We've defined three alternative selection rules: all, subset, and randomized. The subset and randomized versions will pick 1/c rows from the source. The chooser expression will build an iterable over True and False values based on one of the selection rules. The rows to be kept are selected by applying the source iterable to the row selection iterable.
Since all of this is non-strict, rows are not read from the source until required. This allows us to process very large sets of data efficiently. Also, the relative simplicity of the Python code means that we don't really need a complex configuration file and an associated parser to make choices among the selection rules. We have the option to use this bit of Python code as the configuration for a larger data sampling application.
Picking subsets with islice()
In Chapter 4, Working with Collections, we looked at slice notation to select subsets from a collection. Our example was to pair up items sliced from a list object. The following is a simple list:
flat= ['2', '3', '5', '7', '11', '13', '17', '19', '23', '29', '31', '37', '41', '43', '47', '53', '59', '61', '67', '71',... ]
We can create pairs using list slices as follows:
zip(flat[0::2], flat[1::2])
The islice() function gives us similar capabilities without the overhead of materializing a list object, and it looks like the following:
flat_iter_1= iter(flat)
flat_iter_2= iter(flat)
zip(islice(flat_iter_1, 0, None, 2), islice(flat_iter_2, 1, None, 2))
We created two independent iterators over a flat list of data points. These might be two separate iterators over an open file or a database result set. The two iterators need to be independent so that change in one islice() function doesn't interfere with the otherislice() function.
The two sets of arguments to the islice() function are similar to the flat[0::2] and flat[1::2] methods. There's no slice-like shorthand, so the start and stop argument values are required. The step can be omitted and the default value is 1. This will produce a sequence of two tuples from the original sequence:
[(2, 3), (5, 7), (11, 13), (17, 19), (23, 29), ... (7883, 7901), (7907, 7919)]
Since islice() works with an iterable, this kind of design will work with extremely large sets of data. We can use this to pick a subset out of a larger set of data. In addition to using the filter() or compress() functions, we can also use the islice(source,0,None,c)method to pick ![]() items from a larger set of data.
items from a larger set of data.
Stateful filtering with dropwhile() and takewhile()
The dropwhile() and takewhile() functions are stateful filter functions. They start in one mode; the given predicate function is a kind of flip-flop that switches the mode. The dropwhile() function starts in reject mode; when the function becomes False, it switches to pass mode. The takewhile() function starts in pass mode; when the given function becomes False, it switches into reject mode.
Since these are filters, both functions will consume the entire iterable. Given an infinite iterator like the count() function, it will continue indefinitely. Since there's no simple integer overflow in Python, an ill-considered use of dropwhile() or takewhile() functions won't crash after a few billion iterations with integer overflow. It really can run for a very, very long time.
We can use these with file parsing to skip headers or footers in the input. We use the dropwhile() function to reject header rows and pass the remaining data. We use the takewhile() function to pass data and reject trailer rows. We'll return to the simple GPL file format shown in Chapter 3, Functions, Iterators, and Generators. The file has a header that looks as follows:
GIMP Palette
Name: Crayola
Columns: 16
#
This is followed by rows that look like the following example:
255 73 108 Radical Red
We can easily locate the final line of the headers—the # line—using a parser based on the dropwhile() function, as follows:
with open("crayola.gpl") as source:
rdr = csv.reader(source, delimiter='\t')
rows = dropwhile(lambda row: row[0] != '#', rdr)
We created a CSV reader to parse the lines based on tab characters. This will neatly separate the color three tuple from the name. The three tuple will need further parsing. This will produce an iterator that starts with the # line and continues with the rest of the file.
We can use the islice() function to discard the first item of an iterable. We can then parse the color details as follows:
color_rows = islice(rows, 1, None)
colors = ((color.split(), name) for color, name in color_rows)
print(list(colors))
The islice(rows, 1, None) expression is similar to asking for a rows[1:] slice: the first item is quietly discarded. Once the last of the heading rows have been discarded, we can parse the color tuples and return more useful color objects.
For this particular file, we can also use the number of columns located by the CSV reader function. We can use the dropwhile(lambda row: len(row) == 1, rdr) method to discard header rows. This doesn't always work out well in general. Locating the last line of the headers is often easier than trying to locate some general feature that distinguishes header (or trailer) lines from the meaningful file content.
Two approaches to filtering with filterfalse() and filter()
In Chapter 5, Higher-order Functions we looked at the built-in filter() function. The filterfalse() function from the itertools module could be defined from the filter() function, as follows:
filterfalse = lambda pred, iterable:
filter(lambda x: not pred(x), iterable)
As with the filter() function, the predicate function can be of None value. The value of the filter(None, iterable) method is all the True values in the iterable. The value of the filterfalse(None, iterable) method is all of the False values from the iterable:
>>> filter(None, [0, False, 1, 2])
<filter object at 0x101b43a50>
>>> list(_)
[1, 2]
>>> filterfalse(None, [0, False, 1, 2])
<itertools.filterfalse object at 0x101b43a50>
>>> list(_)
[0, False]
The point of having the filterfalse() function is to promote reuse. If we have a succinct function that makes a filter decision, we should be able to use that function to partition input in to pass and reject groups without having to fiddle around with logical negation.
The idea is to execute the following commands:
iter_1, iter_2 = iter(some_source), iter(some_source)
good = filter(test, iter_1)
bad = filterfalse(test, iter_2)
This will obviously include all items from the source. The test() function is unchanged, and we can't introduce a subtle logic bug through improper use of ().
Applying a function to data via starmap() and map()
The built-in map() function is a higher-order function that applies a map() function to items from an iterable. We can think of the simple version of the map() function, as follows:
map(function, arg_iter) == (function(a) for a in arg_iter)
This works well when the arg_iter parameter is a list of individual values. The starmap() function in the itertools module is simply the *a version of the map() function, which is as follows:
starmap(function, arg_iter) == (function(*a) for a in arg_iter)
This reflects a small shift in the semantics of the map() function to properly handle a tuple-of-tuples structure.
The map() function can also accept multiple iterables; the values from these additional iterables are zipped and it behaves like the starmap() function. Each zipped item from the source iterables becomes multiple arguments to the given function.
We can think of the map(function, iter1, iter2, ..., itern) method being defined as the following two commands:
(function(*args) for args in zip(iter1, iter2, ..., itern))
starmap(function, zip(iter1, iter2, ..., itern))
Various iterator values are used to construct a tuple of arguments via the *args construct. In effect, starmap() function is like this more general case. We can build the simple map() function from the more general starmap() function.
When we look at the trip data, from the preceding commands, we can redefine the construction of a Leg object based on the starmap() function. Prior to creating Leg objects, we created pairs of points. Each pair looks as follows:
((Point(latitude=37.54901619777347, longitude=-76.33029518659048), Point(latitude=37.840832, longitude=-76.273834)), ...,(Point(latitude=38.330166, longitude=-76.458504), Point(latitude=38.976334, longitude=-76.473503)))
We could use the starmap() function to assemble the Leg objects, as follows:
with urllib.request.urlopen(url) as source:
path_iter = float_lat_lon(row_iter_kml(source))
pair_iter = legs(path_iter)
make_leg = lambda start, end: Leg(start, end, haversine(start,end))
trip = list(starmap(make_leg, pair_iter))
The legs() function creates pairs of point objects that reflect the start and end of a leg of the voyage. Given these pairs, we can create a simple function, make_leg, which accepts a pair of Points object, and returns a Leg object with the start point, end point, and distance between the two points.
The benefit of the starmap(function, some_list) method is to replace a potentially wordy (function(*args) for args in some_list) generator expression.
Cloning iterators with tee()
The tee() function gives us a way to circumvent one of the important Python rules for working with iterables. The rule is so important, we'll repeat it here.
Note
Iterators can be used only once.
The tee() function allows us to clone an iterator. This seems to free us from having to materialize a sequence so that we can make multiple passes over the data. For example, a simple average for an immense dataset could be written in the following way:
def mean(iterator):
it0, it1= tee(iterator,2)
s0= sum(1 for x in it0)
s1= sum(x for x in it1)
return s0/s1
This would compute an average without appearing to materialize the entire dataset in memory in any form.
While interesting in principle, the tee() function's implementation suffers from a severe limitation. In most Python implementations, the cloning is done by materializing a sequence. While this circumvents the "one time only" rule for small collections, it doesn't work out well for immense collections.
Also, the current implementation of the tee() function consumes the source iterator. It might be nice to create some syntactic sugar to allow unlimited use of an iterator. This is difficult to manage in practice. Instead, Python obliges us to optimize the tee() function carefully.
The itertools recipes
The itertools chapter of the Python library documentation, Itertools Recipes, is outstanding. The basic definitions are followed by a series of recipes that are extremely clear and helpful. Since there's no reason to reproduce these, we'll reference them here. They should be considered as required reading on functional programming in Python.
Note
10.1.2 section, Itertools Recipes of Python Standard Library, is a wonderful resource. See
https://docs.python.org/3/library/itertools.html#itertools-recipes.
It's important to note that these aren't importable functions in the itertools modules. A recipe needs to be read and understood and then, perhaps, copied or modified before inclusion in an application.
The following table summarizes some of the recipes that show functional programming algorithms built from the itertools basics:
|
Function Name |
Arguments |
Results |
|
take |
(n, iterable) |
This returns the first n items of the iterable as a list. This wraps a use of islice() in a simple name. |
|
tabulate |
(function, start=0) |
This returns function(0) and function(1). This is based on a map(function, count()). |
|
consume |
(iterator, n) |
This advances the iterator n steps ahead. If n is None, iterator consumes the steps entirely. |
|
nth |
(iterable, n, default=None) |
This returns the nth item or a default value. This wraps the use of islice() in a simple name. |
|
quantify |
(iterable, pred=bool) |
This counts how many times the predicate is true. This uses sum() and map(), and relies on the way a Boolean predicate is effectively 1 when converted to an integer value. |
|
padnone |
(iterable) |
This returns the sequence elements and then returns None indefinitely. This can create functions that behave like zip_longest() or map(). |
|
ncycles |
(iterable, n) |
This returns the sequence elements n times. |
|
dotproduct |
(vec1, vec2) |
This is the essential definition of a dot product. Multiply two vectors and find the sum of the result. |
|
flatten |
(listOfLists) |
This flattens one level of nesting. This chains the various lists together into a single list. |
|
repeatfunc |
(func, times=None, *args) |
This calls to func repeatedly with specified arguments. |
|
pairwise |
(iterable): |
s -> (s0,s1), (s1,s2), (s2, s3). |
|
grouper |
(iterable, n, fillvalue=None) |
Collect data into fixed length chunks or blocks. |
|
roundrobin |
(*iterables) |
roundrobin('ABC', 'D', 'EF') --> A D E B F C |
|
partition |
(pred, iterable) |
This uses a predicate to partition entries into False entries and True entries. |
|
unique_ everseen |
(iterable, key=None) |
This lists unique elements, preserving order. Remembers all elements ever seen. unique_ everseen('AAAABBBCCDAABBB') - -> A B C D. |
|
unique_ justseen |
(iterable, key=None) |
This lists unique elements, preserving order. Remembers only the element just seen. unique_justseen('AAAABBBCCDAABBB') - -> A B C D A B. |
|
iter_except |
(func, exception, first=None) |
Call a function repeatedly until an exception is raised. This can be used to iterate until KeyError or IndexError. |
Summary
In this chapter, we've looked at a number of functions in the itertools module. This library module provides a number of functions that help us to work with iterators in sophisticated ways.
We've looked at the infinite iterators; these repeat without terminating. These include the count(), cycle(), and repeat() functions. Since they don't terminate, the consuming function must determine when to stop accepting values.
We've also looked at a number of finite iterators. Some of these are built-in and some of these are part of the itertools module. These work with a source iterable, so they terminate when that iterable is exhausted. These functions include enumerate(), accumulate() ,chain() , groupby() , zip_longest(), zip(), compress(), islice(), dropwhile(), takewhile(), filterfalse(), filter(), starmap(), and map(). These functions allow us to replace possibly complex generator expressions with simpler-looking functions.
Additionally, we looked at the recipes from the documentation, which provide yet more functions we can study and copy for our own applications. The recipes list shows a wealth of common design patterns.
In Chapter 9, More Itertools Techniques, we'll continue our study of the itertools module. We'll look at the iterators focused on permutations and combinations. These don't apply to processing large sets of data. They're a different kind of iterator-based tool.