Python and HDF5 (2013)
Chapter 9. Concurrency: Parallel HDF5, Threading, and Multiprocessing
Over the past 10 years or so, parallel code has become crucial to scientific programming. Nearly every modern computer has at least two cores; dedicated workstations are readily available with 12 or more. Plunging hardware prices have made 100-core clusters feasible even for small research groups.
As a rapid development language, and one with easy access to C and FORTRAN libraries, Python is increasingly being used as a top-level “glue” language for such platforms. Scientific programs written in Python can leverage existing “heavy-lifting” libraries written in C or FORTRAN using any number of mechanisms, from ctypes to Cython to the built-in NumPy routines.
This chapter discusses the various mechanisms in Python for writing parallel code, and how they interact with HDF5.
Python Parallel Basics
Broadly speaking, there are three ways to do concurrent programming in Python: threads, the multiprocessing module, and finally by using bindings for the Message Passing Interface (MPI).
Thread-based code is fine for GUIs and applications that call into external libraries that don’t tie up the Python interpreter. As we’ll see in a moment, you can’t use more than one core’s worth of time when running a pure-Python program. There’s also no performance advantage on the HDF5 side to using threads, since the HDF5 library serializes all calls.
multiprocessing is a more recent built-in module available with Python, which provides support for basic fork()-based parallel processing. The main restriction is that your parallel processes can’t share a single HDF5 file, even if the file is opened read-only. This is a limitation of the HDF5 library. multiprocessing-based code is great for long-running, CPU-bound problems in which data I/O is a relatively small component handled by the master process. It’s the simplest way in Python to write parallel code that uses more than one core, and strongly recommended for general-purpose computation.
For anything else, MPI-based Parallel HDF5 is by far the best way to go. MPI is the official “flavor” of parallelism supported by the HDF5 library. You can have an unlimited number of processes, all of which share the same open HDF5 file. All of them can read and write data, and modify the file’s structure. Programs written this way require a little more care, but it’s the most elegant and highest-performance way to use HDF5 in a parallel context.
By the way, there’s even a fourth mechanism for parallel computing, which is becoming increasingly popular. IPython, which you’re probably already using as a more convenient interpreter interface, has its own clustering capabilities designed around the ZeroMQ networking system. It can even use MPI on the back end to improve the performance of parallel code. At the moment, not much real-world use of h5py in the IPython parallel model is known to the author. But you should definitely keep an eye on it, if for no other reason than the very convenient interface that IPython provides for clustering.
Threading
HDF5 has no native support for thread-level parallelism. While you can safely use HDF5 (and h5py) from threaded programs, there will be no performance advantage. But for a lot of applications, that’s not a problem. GUI apps, for example, spend most of their time waiting for user input.
Before we start, it’s important to review a few basics about threaded programming in the Python world. Python itself includes a single master lock that governs access to the interpreter’s functions, called the Global Interpreter Lock or GIL. This lock serializes access from multiple threads to basic resources like object reference counting. You can have as many threads as you like in a Python program, but only one at a time can use the interpreter.
This isn’t such a big deal, particularly when writing programs like GUIs or web-based applications that spend most of their time waiting for events. In these situations, the GIL is “released” and other threads can use the interpreter while an I/O-bound thread is waiting.
h5py uses a similar concept. Access to HDF5 is serialized using locking, so only one thread at a time can work with the library. Unlike other I/O mechanisms in Python, all use of the HDF5 library is “blocking”; once a call is made to HDF5, the GIL is not released until it completes.
Figure 9-1 shows schematically how this works. If you have multiple threads running, if one of them calls into HDF5 (for example, to write a large dataset to disk), the others will not proceed until the call completes.
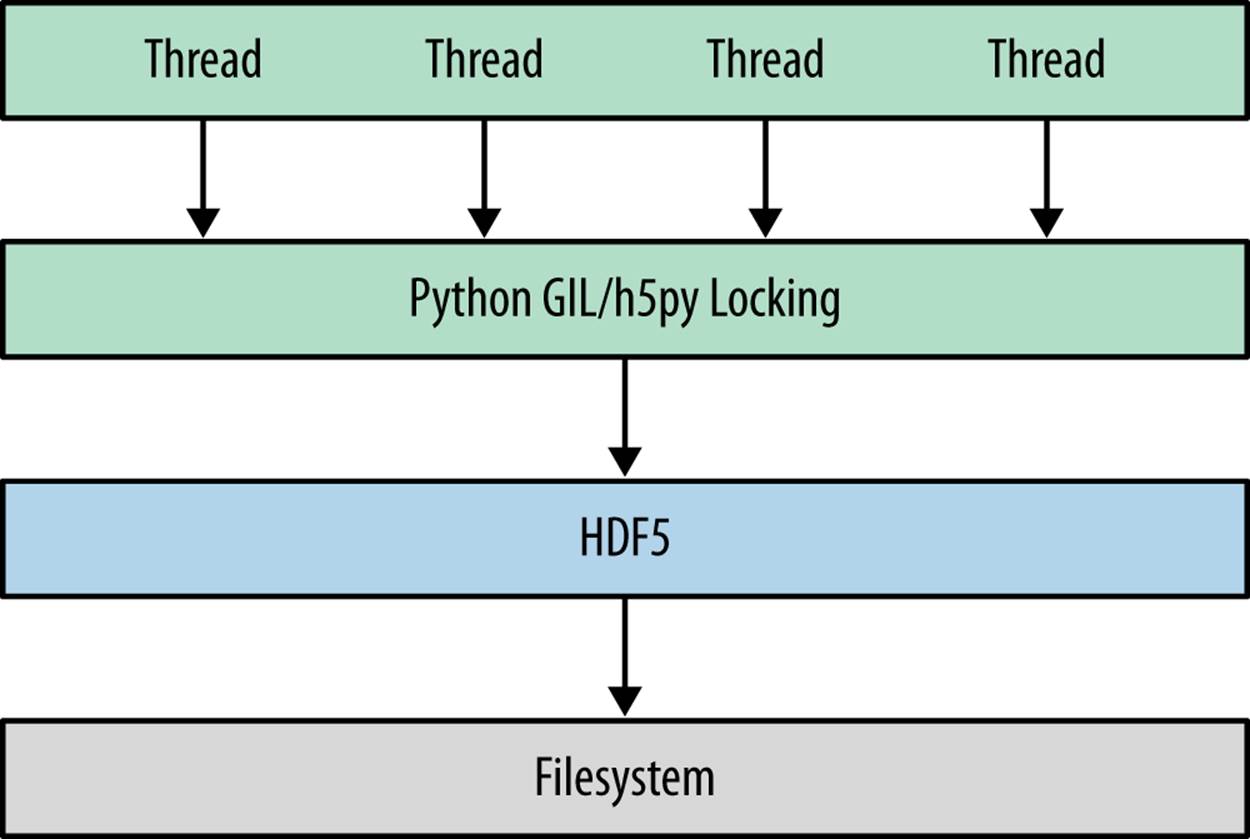
Figure 9-1. Outline of a threading-based program using HDF5
The h5py package is “thread-safe,” in that you can safely share objects between threads without corruption, and there’s no global state that lets one thread stomp on another. However, certain high-level operations are not yet guaranteed to be atomic. Therefore, it’s recommended that you manage access to your HDF5 objects by using recursive locks.
Here’s an example: we’ll create a single shared HDF5 file and two threads that do some computation and write to it. Access to the file is managed using an instance of the threading.RLock class:
import threading
import time
import random
import numpy as np
import h5py
f = h5py.File("thread_demo.hdf5", "w")
dset = f.create_dataset("data", (2, 1024), dtype='f')
lock = threading.RLock()
class ComputeThread(threading.Thread):
def __init__(self, axis):
self.axis = axis # One thread does dset[0,:], the other dset[1, :].
threading.Thread.__init__(self)
def run(self):
""" Perform a series of (simulated) computations and save to dataset.
"""
for idx inxrange(1024):
random_number = random.random()*0.01
time.sleep(random_number) # Perform computation
with lock:
dset[self.axis, idx] = random_number # Save to dataset
thread1 = ComputeThread(0)
thread2 = ComputeThread(1)
thread1.start()
thread2.start()
# Wait until both threads have finished
thread1.join()
thread2.join()
f.close()
NOTE
There’s also something out there called “thread-safe” HDF5. This is a compile-time option for the HDF5 library itself. Using it won’t hurt anything, but it’s not required for thread-safe use of h5py.
Multiprocessing
Because of the GIL, a threading-based Python program can never use more than the equivalent of one processor’s time. This is an annoyance when it comes to writing native-Python programs for parallel processing. Historically, people have used ad hoc solutions involving multiple instances of Python, communicating through the filesystem.
Since version 2.6, Python has included an “entry-level” parallel-processing package called multiprocessing. It provides a threading-like way to manage multiple instances of Python.
It is possible to use multiprocessing with HDF5, provided some precautions are taken. The most important thing to remember is that new processes inherit the state of the HDF5 library from the parent process. It’s very easy to end up in a situation where multiple processes end up “fighting” each other over an open file. At the time of writing (October 2013), this is even true for files that are opened read-only. This has to do with the gory details of how multiprocessing is implemented, using the fork() system call on Linux/Unix operating systems. The HDF Group is aware of this limitation; you should check the most recent documentation for h5py/PyTables to see if it has been resolved.
In the meantime, to avoid problems, here are some things to try:
1. Do all your file I/O in the main process, but don’t have files open when you invoke the multiprocessing features.
2. Multiple subprocesses can safely read from the same file, but only open it once the new process has been created.
3. Have each subprocess write to a different file, and merge them when finished.
Figure 9-2 shows workflow (1). The initial process is responsible for file I/O, and communicates with the subprocesses through queues and other multiprocessing constructs.
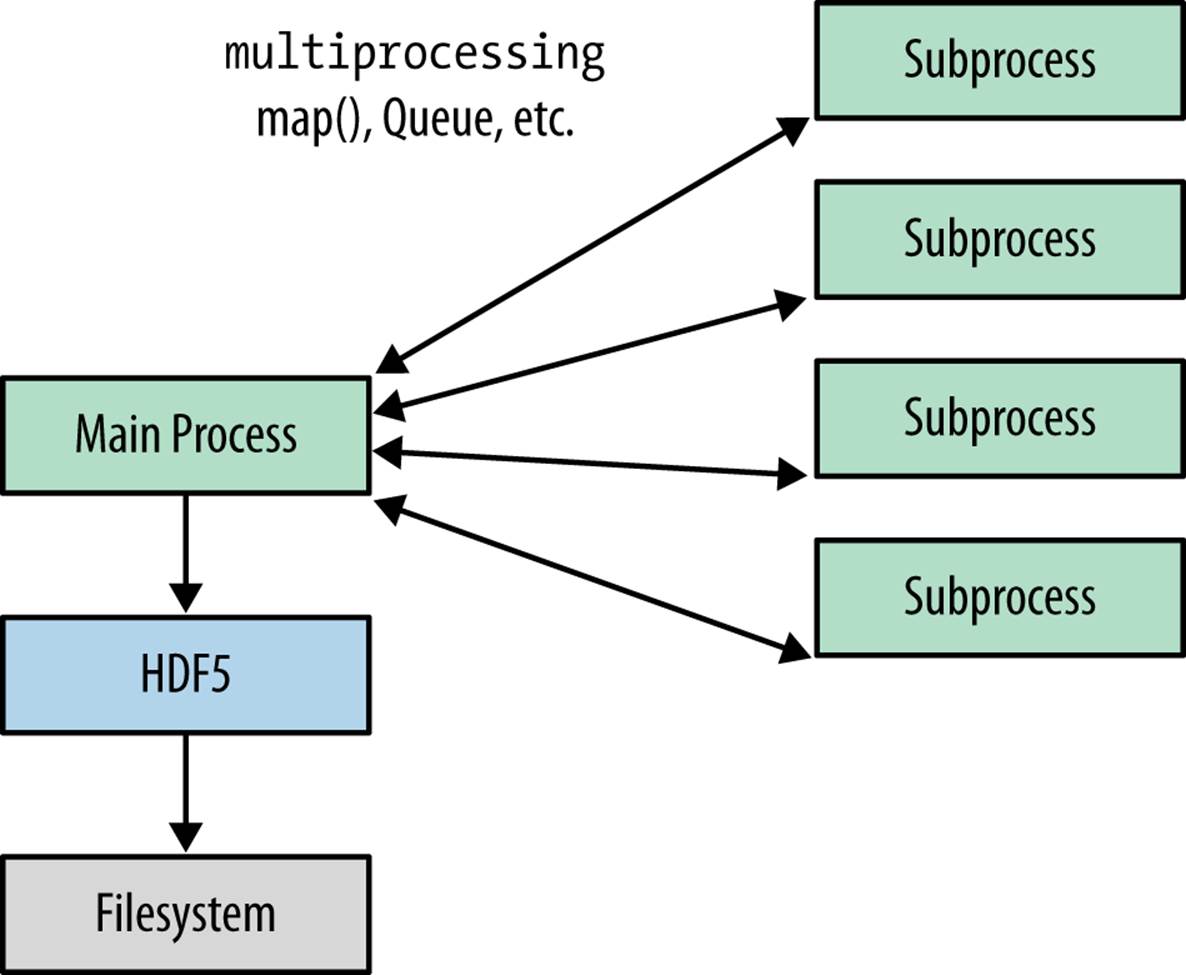
Figure 9-2. Multiprocessing-based approach to using HDF5
One mechanism for “Pythonic” parallel computation is to use “process pools” that distribute the work among worker processes. These are instances of multiprocessing.Pool, which among other things have a parallel equivalent of the built-in map():
>>> import string
>>> from multiprocessing import Pool
>>> p = Pool(2) # Create a 2-process pool
>>> words_in = ['hello', 'some', 'words']
>>> words_out = p.map(string.upper, words_in)
>>> print words_out
['HELLO', 'SOME', 'WORDS']
Here’s an example of using HDF5 with Pool. Suppose we had a file containing a 1D dataset of coordinate pairs, and we wanted to compute their distance from the origin. This is the kind of problem that is trivial to parallelize, since each computation doesn’t depend on the others. First, we create our file, containing a single coords dataset:
with h5py.File('coords.hdf5', 'w') as f:
dset = f.create_dataset('coords', (1000, 2), dtype='f4')
dset[...] = np.random.random((1000,2))
Our program will use a simple one-liner for the distance measurement, and a four-process Pool to carry out the 1,000 conversions required. Note that we don’t have any files open when invoking map:
import numpy as np
from multiprocessing import Pool
import h5py
def distance(arr):
""" Compute distance from origin to the point (arr is a shape-(2,) array)
"""
return np.sqrt(np.sum(arr**2))
# Load data and close the input file
with h5py.File('coords.hdf5', 'r') as f:
data = f['coords'][...]
# Create a 4-process pool
p = Pool(4)
# Carry out parallel computation
result = np.array(p.map(distance, data))
# Write the result into a new dataset in the file
with h5py.File('coords.hdf5') as f:
f['distances'] = result
Doing anything more complex with multiprocessing and HDF5 gets complicated. Your processes can’t all access the same file. Either you do your I/O explicitly in the main process (as shown), or you have each process generate a bunch of smaller “shard” files and join them together when you’re done:
import os
import numpy as np
from multiprocessing import Pool
import h5py
def distance_block(idx):
""" Read a 100-element coordinates block, compute distances, and write
back out again to a process-specific file.
"""
with h5py.File('coords.hdf5','r') as f:
data = f['coords'][idx:idx+100]
result = np.sqrt(np.sum(data**2, axis=1))
with h5py.File('result_index_%d.hdf5'%idx, 'w') as f:
f['result'] = result
# Create out pool and carry out the computation
p = Pool(4)
p.map(distance_block, xrange(0, 1000, 100))
with h5py.File('coords.hdf5') as f:
dset = f.create_dataset('distances', (1000,), dtype='f4')
# Loop over our 100-element "chunks" and merge the data into coords.hdf5
for idx inxrange(0, 1000, 100):
filename = 'result_index_%d.hdf5'%idx
with h5py.File(filename, 'r') as f2:
data = f2['result'][...]
dset[idx:idx+100] = data
os.unlink(filename) # no longer needed
That looks positively exhausting, mainly because of the limitations on passing open files to child processes. What if there were a way to share a single file between processes, automatically synchronizing reads and writes? It turns out there is: Parallel HDF5.
MPI and Parallel HDF5
Figure 9-3 shows how an application works using Parallel HDF5, in contrast to the threading and multiprocessing approaches earlier. MPI-based applications work by launching multiple parallel instances of the Python interpreter. Those instances communicate with each other via the MPI library. The key difference compared to multiprocessing is that the processes are peers, unlike the child processes used for the Pool objects we saw earlier.
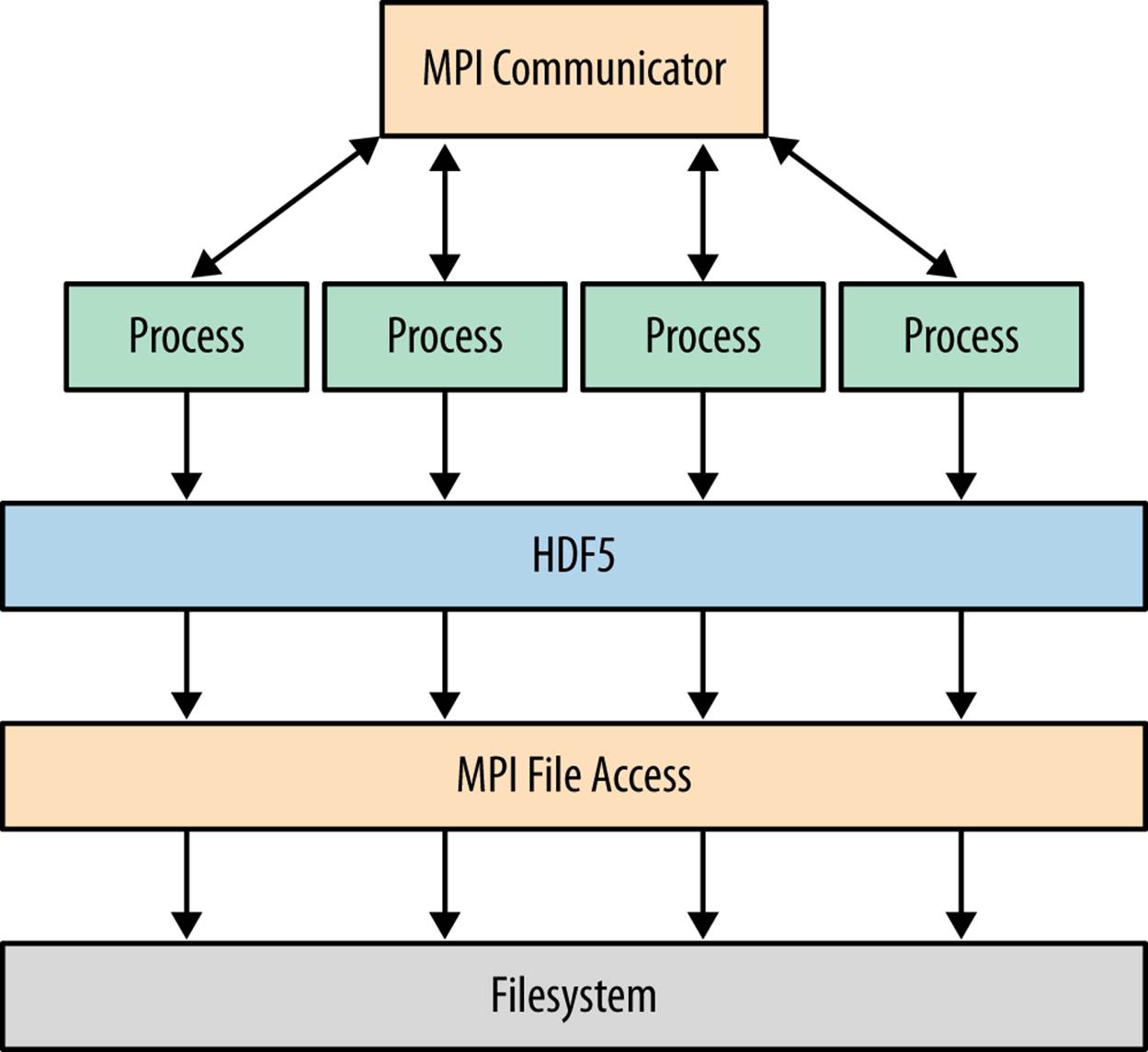
Figure 9-3. MPI-based approach using Parallel HDF5
Of course, this also means that all file access has to be coordinated though the MPI library as well. If not, multiple processes would “fight” over the same file on disk. Thankfully, HDF5 itself handles nearly all the details involved with this. All you need to do is open shared files with a special driver, and follow some constraints for data consistency.
Let’s take a look at how MPI-based programs work in Python before diving into Parallel HDF5.
A Very Quick Introduction to MPI
MPI-based programs work a little differently from thread-based or multiprocessing programs. Since there’s no explicit creation of processes on the Python side, you write a Python script and launch it with the special program mpiexec. Here’s the “Hello World” example for MPI:
from mpi4py import MPI
comm = MPI.COMM_WORLD
print "Hello World (from process %d)" % comm.rank
To run it with four processes, we launch Python via mpiexec:
$ mpiexec -n 4 python demo.py
Hello World (from process 0)
Hello World (from process 1)
Hello World (from process 3)
Hello World (from process 2)
The COMM_WORLD object is an MPI communicator. Communication between processes all happens though the MPI library, via these objects. In addition to “send” and “receive” methods, communicators in MPI support a variety of advanced “scatter” operations to distribute work among processes. Since a complete treatment of MPI features is beyond the scope of this book, we will limit ourselves to a few basic features.
The COMM_WORLD attribute rank is an integer that identifies the running process. You’ll notice that the output statements print “out of order” with respect to rank. As with threads, since our four processes run in parallel there’s no guarantee as to which will finish first. Later on we’ll examine some basic methods for interprocess synchronization.
NOTE
Check out the mpi4py website for a great introduction to using MPI from Python, and a large number of examples.
MPI-Based HDF5 Program
Building h5py in MPI mode is simple. The tricky bit is getting access to a version of HDF5 compiled in “parallel” mode; increasingly, this is available as a built-in package on most Linux distributions. Then, when you build h5py, just enable the --mpi option:
$ python setup.py build --mpi [--hdf5=/path/to/parallel/hdf5]
$ [sudo] python setup.py install
NOTE
If you have installation problems, consult the guide at www.h5py.org for tips and additional examples. The examples shown here use an OpenMPI build of HDF5, running on Linux.
On the Python side, all you have to do is open your file with the mpio driver. You have to supply an MPI communicator as well; COMM_WORLD does nicely:
from mpi4py import MPI
import h5py
f = h5py.File("foo.hdf5", "w", driver="mpio", comm=MPI.COMM_WORLD)
In contrast to the other file drivers, a file opened with mpio can safely be accessed from multiple processes.
Here’s the MPI equivalent of our distance-calculation code. Note that the division of work is done implicitly (in this case using the process rank) in contrast to the explicit map()-based approach we saw in the multiprocessing example:
import numpy as np
import h5py
from mpi4py import MPI
comm = MPI.COMM_WORLD # Communicator which links all our processes together
rank = comm.rank # Number which identifies this process. Since we'll
# have 4 processes, this will be in the range 0-3.
f = h5py.File('coords.hdf5', driver='mpio', comm=comm)
coords_dset = f['coords']
distances_dset = f.create_dataset('distances', (1000,), dtype='f4')
idx = rank*250 # This will be our starting index. Rank 0 handles coordinate
# pairs 0-249, Rank 1 handles 250-499, Rank 2 500-749, and
# Rank 3 handles 750-999.
coords = coords_dset[idx:idx+250] # Load process-specific data
result = np.sqrt(np.sum(coords**2, axis=1)) # Compute distances
distances_dset[idx:idx+250] = result # Write process-specific data
f.close()
Collective Versus Independent Operations
MPI has two flavors of operation: collective, which means that all processes have to participate (and in the same order), and independent, which means each process can perform the operation (or not) whenever and in whatever order it pleases.
With HDF5, the main requirement is this: modifications to file metadata must be done collectively. Here are some things that qualify:
§ Opening or closing a file
§ Creating or deleting new datasets, groups, attributes, or named types
§ Changing a dataset’s shape
§ Moving or copying objects in the file
Generally this isn’t a big deal. What it means for your code is the following: when you’re executing different code paths depending on the process rank (or as the result of an interprocess communication), make sure you stick to data I/O only. In contrast to metadata operations, data operations (meaning reading from and writing to existing HDF5) are OK for processes to perform independently.
Here are some simple examples:
from mpi4py import MPI
import h5py
comm = MPI.COMM_WORLD
rank = comm.rank
f = h5py.File('collective_test.hdf5', 'w', driver='mpio', comm=comm)
# RIGHT: All processes participate when creating an object
dset = f.create_dataset('x', (100,), 'i')
# WRONG: Only one process participating in a metadata operation
if rank == 0:
dset.attrs['title'] = "Hello"
# RIGHT: Data I/O can be independent
if rank == 0:
dset[0] = 42
# WRONG: All processes must participate in the same order
if rank == 0:
f.attrs['a'] = 10
f.attrs['b'] = 20
else:
f.attrs['b'] = 20
f.attrs['a'] = 10
When you violate this requirement, generally you won’t get an exception; instead, various Bad Things will happen behind the scenes, possibly endangering your data.
Note that “collective” does not mean “synchronized.” Although all processes in the preceding example call create_dataset, for example, they don’t pause until the others catch up. The only requirements are that every process has to make the call, and in the same order.
Atomicity Gotchas
Sometimes, it’s necessary to synchronize the state of multiple processes. For example, you might want to ensure that the first stage of a distributed calculation is finished before moving on to the next part. MPI provides a number of mechanisms to deal with this. The simplest is called “barrier synchronization”—from the Python side, this is simply a function called Barrier that blocks until every process has reached the same point in the program.
Here’s an example. This program generally prints “A” and “B” statements out of order:
from random import random
from time import sleep
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.rank
sleep(random()*5)
print "A (rank %d)" % rank
sleep(random()*5)
print "B (rank %d)" % rank
Running it, we get:
$ mpiexec -n 4 python demo2.py
A (rank 2)
B (rank 2)
A (rank 1)
A (rank 0)
B (rank 1)
A (rank 3)
B (rank 3)
B (rank 0)
Our COMM_WORLD communicator includes a Barrier function. If we add a barrier for all processes just before the “B” print statement, we get:
from random import random
from time import sleep
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.rank
sleep(random()*5)
print "A (rank %d)" % rank
comm.Barrier() # Blocks until all processes catch up
sleep(random()*5)
print "B (rank %d)" % rank
$ mpiexec -n 4 python demo3.py
A (rank 2)
A (rank 3)
A (rank 0)
A (rank 1)
B (rank 2)
B (rank 0)
B (rank 1)
B (rank 3)
Now that you know about Barrier, what do you think the following two-process program outputs?
import h5py
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.rank
with h5py.File('atomicdemo.hdf5', 'w', driver='mpio', comm=comm) as f:
dset = f.create_dataset('x', (1,), dtype='i')
if rank == 0:
dset[0] = 42
comm.Barrier()
if rank == 1:
print dset[0]
If you answered “42,” you’re wrong. You might get 42, and you might get 0. This is one of the most irritating things about MPI from a consistency standpoint. The default write semantics do not guarantee that writes will have completed before Barrier returns and the program moves on. Why? Performance. Since MPI is typically used for huge, thousand-processor problems, people are willing to put up with relaxed consistency requirements to get every last bit of speed possible.
Starting with HDF5 1.8.9, there is a feature to get around this. You can enable MPI “atomic” mode for your file. This turns on a low-level feature that trades performance for strict consistency requirements. Among other things, it means that Barrier (and other MPI synchronization methods) interact with writes the way you expect. This modified program will always print “42”:
import h5py
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.rank
with h5py.File('atomicdemo.hdf5', 'w', driver='mpio', comm=comm) as f:
f.atomic = True # Enables strict atomic mode (requires HDF5 1.8.9+)
dset = f.create_dataset('x', (1,), dtype='i')
if rank == 0:
dset[0] = 42
comm.Barrier()
if rank == 1:
print dset[0]
The trade-off, of course, is reduced performance. Generally the best solution is to avoid passing data from process to process through the file. MPI has great interprocess communication tools. Use them!