Emerging Trends in Image Processing, Computer Vision, and Pattern Recognition, 1st Edition (2015)
Part II. Computer Vision and Recognition Systems
Chapter 25. Biometric analysis for finger vein data
Two-dimensional kernel principal component analysis
Sepehr Damavandinejadmonfared; Vijay Varadharajan Department of Computing, Advanced Cyber Security Research Centre, Macquarie University, Sydney, New South Wales, Australia
Abstract
In this paper, a whole identification system is introduced for finger vein recognition. The proposed algorithm first maps the input data into kernel space, then; two-dimensional principal component analysis (2DPCA) is applied to extract the most valuable features from the mapped data. Finally, Euclidian distance classifies the features and the final decision is made. Because of the natural shape of human fingers, the image matrixes are not square, which makes them possible to use kernel mappings in two different ways—along row or column directions. Although some research has been done on the row and column direction through 2DPCA, our argument is how to map the input data in different directions and get a square matrix out of it to be analyzed by 2DPCA. In this research, we have explored this area in details and obtained the most significant way of mapping finger vein data which results in consuming the least time and achieving the highest accuracy for finger vein identification system. The authenticity of the results and the relationship between the finger vein data and our contribution are also discussed and explained. Furthermore, extensive experiments were conducted to prove the merit of the proposed system.
Keywords
Biometrics
Finger vein recognition
2D Principal component analysis
Kernel principal component analysis
1 Introduction
Traditionally, private information is considered as passwords and personal identification numbers (PINs) among the society, which is easy to use but vulnerable to the risk of exposure and being stolen or forgotten. Biometrics [1,2], however, has been attracting researchers' and industry's attention more and more as it is believed that biometrics is a promising alternative to the traditionally used password or PIN-based authentication techniques [3]. Nowadays, there are several different biometrics systems under research such as face recognition, finger print, palm print, voice recognition, iris recognition, and so on [1,4–7]. There are two main challenges in terms of biometrics systems. The first one is that the main element by which the identity is verified or identified is accessible and forgeable, and the second one is that the rate of reliability of the mentioned systems in terms of having a satisfactory accuracy rate is not acceptable. For instance, finger and palm prints are usually frayed; iris images and voice signature are easily forged; face recognition could be considered difficult and unreliable when there are occlusions or face-lifts. Finger vein recognition [8–11], however, is more secure and convenient and has none of the mentioned drawbacks because of the following three reasons: (1) human veins are mostly invisible and located inside the body; therefore, it is difficult to be forged or stolen, (2) it is more acceptable for the user as capturing finger vein images is noninvasive and contactless, and (3) the finger vein data can only be captured from a live individual. It is thus a convincing proof that the subject whose finger vein [12] is successfully captured is alive.
Because the data in finger vein recognition are “image,” there are several methods for analyzing and classifying the images in such a recognition system. Principal component analysis (PCA) [3,13] is one of the common and powerful methods of pattern recognition and feature extraction which has been used a lot in biometrics. There have been several improvements on PCA such as kernel PCA (KPCA) [13,14], and kernel entropy PCA (KECA) [15–17] so far. See the book [18] for kernel methods in pattern analysis. One-dimensional (1D) KPCA [19] and KECA [20,21] have been proposed for finger vein recognition which led to promising results [22]. The main drawback of 1D PCA, however, is that after converting the 2D matrix to 1D, the dimension of the data is too high which results in having a very time-consuming and even inaccurate system. A highlighted improvement on 1DPCA is 2DPCA [23–26] in which the image matrix is not converted to 1D. This method has two main advantages over 1DPCA which are being much faster, and having higher accuracy. After proposing 2DPCA, kernel 2DPCA [25] was introduced in which the data are first mapped to another space using different kernel methods and then 2DPCA is implemented on the mapped data. It is believed that by transforming the data into the appropriate space first and applying 2DPCA on the mapped data, the accuracy rate will have a dramatic increase. This work is an extension of Ref. [27] presented in IPCV 2014 conference. As 2DPCA is applied on 2D image matrixes directly, there has been a great amount of research on the direction of the analysis. 2DPCA can be applied in row direction, column direction, or both. In this paper, however, different directions of the matrixes for mapping the data into kernel space are argued. We chose kernel polynomial degree one as mapping function and applied input images in row and column directions. The direction of mapping is important in our system because 2DPCA is applied on the mapped data and extracts as many eigenvectors as the dimension of the mapped data, meaning that if we have an (N × N) matrix, we will have N eigenvectors and their corresponding eigenvalues. For example, assuming our input matrixes are 60 × 180, applying the mapping function in row and column directions will result in 60 × 60 and 180 × 180 matrixes, respectively. Applying 2DPCA on 60 × 60 or 180 × 180 matrixes will lead to a great difference in accuracies and time duration for the system.
The remaining of this paper is organized as follows: in Section 2, image acquisition is explained. In Section 3, 2D principal component is introduced briefly. In Section 4, kernel mapping along row and column direction is explained. In Section 5, finger vein recognition algorithm is proposed. In Section 6, experimental results on the finger vein database are discussed. Finally, Section 7 concludes the paper.
2 Image Acquisition
In this section, we briefly explain how the finger vein images were captured and what devices were used building the scanner. Based on the proven scientific fact that the light rays can be absorbed by deoxygenated hemoglobin in the vein, absorption coefficient of the vein is higher than other parts of finger. Having said that we designed a scanner consisting of four low-cost prototype devices such as infrared LEDs (830 nm wavelength) and the control circuit to drive the LEDs properly, a camera to capture the images, an infrared pass filter and a computer to process the images. To make the camera sensitive to the IR light, there have been some modifications to it; the IR blocking filter was removed and replaced with an IR pass filter which blocks visible wavelengths of light and passes the IR light (Figure 1).
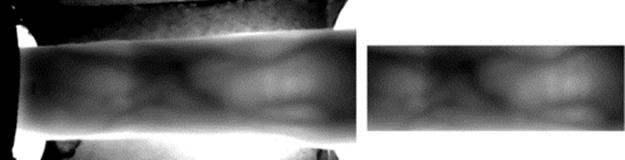
FIGURE 1 A subset of samples captured from a subject.
3 Two-dimensional principal component analysis
The main idea of two-dimensional principal component analysis (2DPCA) is to project the image A, which is represented as a random m × n matrix, onto X that is an n-dimensional unitary column vector:
![]() (1)
(1)
Therefore, the projected feature vector of image A is achieved from Equation (1). Finding the appropriate projection vector (X) is the goal. To evaluate the discriminatory power of X, the total scatter of projected samples can be used, which could be characterized by tracing the covariance matrix of projected features vectors. The following criterion is introduced from this point of view:
![]() (2)
(2)
where Sx represents the covariance matrix of the projected feature vector and tr(Sx) is the trace of Sx. To maximize the criterion in Equation (2), the appropriate projection direction X needs to be found. Sx is introduced by
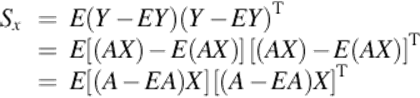
Therefore,
![]() (3)
(3)
Defining the image covariance scatter matrix, we have
![]() (4)
(4)
We now obtain the n × n matrix of Gt from all training images, where there are m training images of size m × n. Thus, Gt can be evaluated by
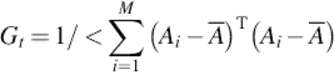 (5)
(5)
where ![]() is the mean matrix of input images and finally we have
is the mean matrix of input images and finally we have
![]() (6)
(6)
First, the n × n matrix of Gt is calculated from all of the training images. Then, the unitary vectors X are obtained by getting the eigenvector matrix of Gt. This stage decides how many eigenvectors are to be used in the projection of data. To achieve this, the eigenvalues of the corresponding eigenvectors are arranged in a descending order, and a subset of the higher values is selected. Assuming d eigenvectors (with optimal projection axes X1, X2, …, Xd) are selected, then how to achieve feature extraction and classification stages are explained in the next section.
4 Kernel mapping along row and column direction
4.1 Two-Dimensional KPCA
The main idea of using kernel function in PCA is that the data are first mapped into another space using a mapping function and then PCA is performed on the nonlinearly mapped data. 2DPCA is better than 1D PCA in terms of speed and accuracy. The idea of using kernel function in 2DPCA is to improve the accuracy of the system. With N input images, let Ai be the ith image, where i = 1, 2, …, N, and Aij be the jth row of the matrix Ai, where j = 1, 2, …, n. The nonlinear mapping is defined as follows:
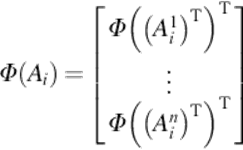 (7)
(7)
The total scatter matrix in kernel 2DPCA can be calculated as
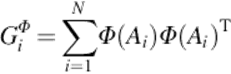 (8)
(8)
Thus,
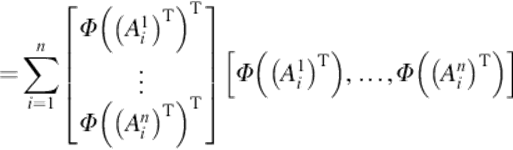
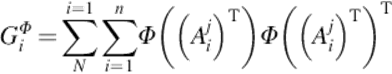 (9)
(9)
In kernel 2DPCA, after achieving Bi = [Y1i, Y1i, …, Ydi] and GtΦ, obtaining projecting axes and the projection and classification procedures are same as in 2DPCA.
4.2 Kernel Mapping in Row and Column Directions and 2DPCA
Equations (7)–(9) demonstrate how the kernel mapping is performed on the input data. Our argument here is that by applying this mapping in different directions (along row and column directions), we will end up having two different data having different dimensions. To further elaborate this, let us assume we have m × n input matrixes (n > m). By applying the kernel mapping function along the row direction (we have m rows meaning that there are m elements for the kernel matrix to be made of), the kernel matrix will be (m × m). In this case, by applying kernel mapping, we reduce the number of input data and its dimension in kernel space which is (n × n). However, if the kernel function is applied along column direction, the kernel matrix will be (n × n). In this case, we have expanded the input data to a higher dimensional space and we have more information to analyze by 2DPCA. Figure 2 shows the detailed diagram of the kernel mapping and 2DPCA on the mapped data in two different directions. It is observed from the diagram that by applying the kernel function from row or column direction, the kernel matrix (K) is squared and with dimension of n or m.
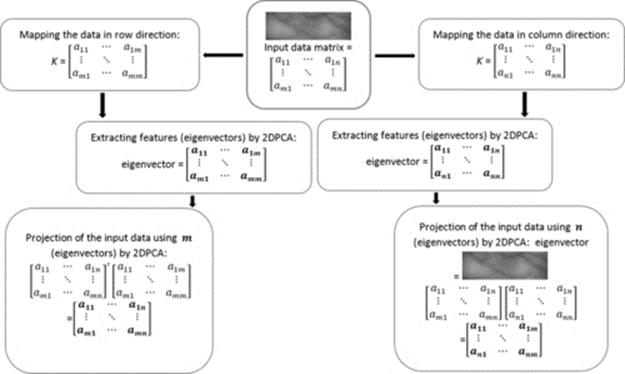
FIGURE 2 Flow diagram of kernel mapping along row and column direction and applying 2DPCA.
This argument is indispensable because the dimension of the data affects the output of the 2DPCA greatly. Having higher dimension and more information and features does not guarantee ending up more promising results and higher accuracies. Furthermore, the higher the dimension, the more time consuming the system is. On the other hand, there has to be a balance between the dimension of the data, the number of used features, and the algorithm that is used to analyze the data.
5 Finger Vein Recognition Algorithm
Our proposed finger vein recognition algorithm is explained in this section. As it is shown in Figure 3, the algorithm consists of five steps; first step is to extract the region of interest (ROI). Second one is to normalize the images. Third step is to map the data into kernel space along the row and column directions which was explained in Section 4. In the fourth step, 2DPCA is applied on the data and features are extracted. Last step is to classify the data using Euclidian distance. The flow diagram of the proposed algorithm is indicated in Figure 3. All steps except for steps 3 and 4, which were explained in Section 4, are introduced in the succeeding part of this section.
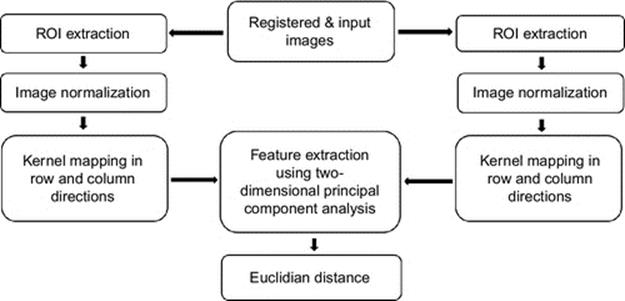
FIGURE 3 Flow diagram of the proposed algorithm.
5.1 ROI Extraction
The unwanted black area around the images should be cropped as this area reduces the accuracy and is considered as nothing but noise. To crop images optimally, the used algorithm consists of three major steps. First of all, the edge is detected. Using the detected edges two horizontal lines are determined and the image is cropped horizontally according to the detected lines. Last but not least, the image is cropped vertically at 5% from the left border and 15% from the right border.
5.2 Image Normalization
In order to achieve the highest accuracy in least time, images are normalized to smaller size after ROI extraction. It is obvious that the smaller the size, the faster the system is. However, if the size of the image is too small, it may cause too much loss of information as well. Therefore, there has to be a balance between size of the images and the accuracy of the system. Based on our experiments, when using 2DPCA to extract the features, the optimal size of finger vein images resulting in both least time consumption and highest accuracy is 20 × 60. Thus, all images are normalized into 20 × 60.
5.3 Feature Extraction and Classification Method
As it was mentioned before, Euclidian distance is used as a classifier in this system. Euclidian distance is a very fast method which, we believe, is appropriate for this system because after using kernel map and 2DPCA, the dimension of the data is reduced and therefore the Euclidian distance is sufficient to be used.
Given an image sample A and the optimal projection axes (selected eigenvectors, X1, X2, …, Xd), the projection will be as follows:
![]() (10)
(10)
Using d axes to project the data onto, we will get d projected feature vectors Y1Y2, …, Yd. These vectors are the principal component of the sample image A. Putting these vectors in the form of a matrix, we will get feature matrix of the image A, which is m × d,B = [Y1, …, Yd].
Then, a nearest neighbor classifier is used to classify the data after transferring all images by 2DPCA and obtaining the feature matrix of them. Considering Bi = [Y1i, Y1i, …, Ydi] and Bj = [Y1i, Y1i, …, Ydi], the Euclidian distance between them is defined as follows:
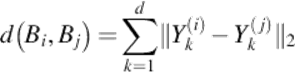 (11)
(11)
6 Experimental results on finger vein database
In this section, the experiments conducted on finger vein data are given and explained. Experimental results are explained in two subsections; column direction analysis in Section 6.1 and row direction analysis in Section 6.2. Our database consists of 10 samples for each of 100 individuals which results in a total number of 1000 images. In each different part of the experiments, three different types of training and testing were used. The size of image samples also varies as well as the number of testing and training. We used two different sizes ([20 × 60] and [10 × 20]) in our experimental section. 2, 3, and 4 random selected images were used to train each time and, respectively, the remaining 8, 7, and 6 images to test. All implementations in each part were repeated as many times as the number of total eigenvectors.
6.1 Experimental Setup-1
In this section, we explain the analysis with respect to the images with size of (20 × 60) for the sake of understanding. To analyze the system in column direction and get the output, we first restate that the images are in dimension of (20 × 60) meaning that if we map them in column direction, there will be 60 samples with the dimension of 20 to map. The output of such a mapping function will be a matrix with the dimension of (60 × 60). By applying 2DPCA on this matrix, there will be 60 eigenvectors extracted with the dimension of 20. In this step, there are 60 different dimensions which could be reduced using projection in 2DPCA meaning that there are 60 different projections using different number of eigenvectors. We conducted the algorithm 60 times in each of three different types (2, 3, and 4 samples to train and 8, 7, and 6 samples to test, respectively) of our implementation and calculated the accuracy rate in each point. The obtained results were then gathered and shown in Figure 4. As it was expected, by adding the number of samples for train, the accuracy goes up no matter which method to use. Another expectation was that by using more eigenvectors, the accuracy rate goes higher up to its optimized point, which here is almost near the dimension of 20. It is observed from Figure 4that using 2, 3, and 4 images to train leads to the accuracy rate of around 90%, 95%, and 97%, respectively. Another prime issue is that the time consumed for this experiment is much more than the next as there are 60 and 20 eigenvectors in column direction mapping. It is also observed that by making the image size smaller the accuracy rate drops. It is because by making the size of images smaller, some information are lost.
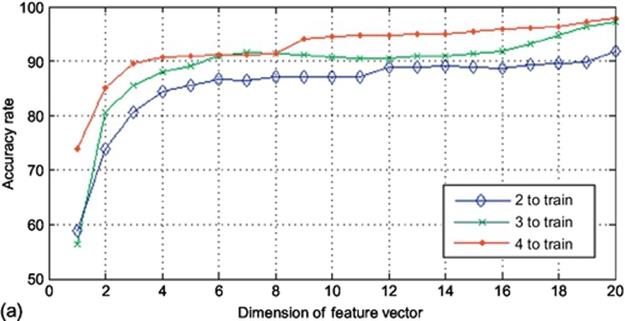
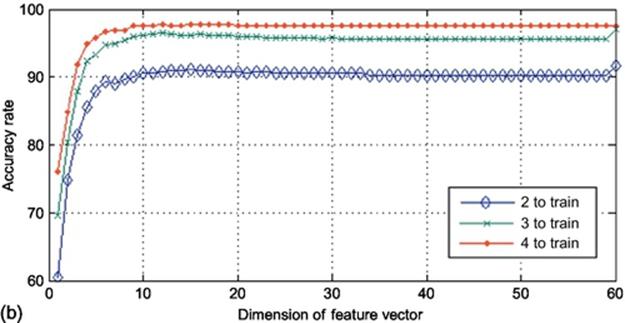
FIGURE 4 Accuracy rates obtained using K2DPCA in column direction on finger vein database: (a) (10 × 20) sample size and (b) (20 × 60) sample size.
6.2 Experimental Setup-2
To analyze the system in row direction, the input images were used in a way that each image consists of 20 samples with the dimension of 60 to map. The output of such a mapping function will be a (20 × 20) matrix. By applying 2DPCA on this matrix, there will be 20 eigenvectors extracted with the dimension of 20. In this step, there are 20 different dimensions which could be reduced using projection in 2DPCA meaning that there are 20 projection manners using different number of eigenvectors. Figure 5 demonstrates the accuracy rate of the experiments along the row direction. Implementing this method using 2, 3, and 4 images to train results to the accuracy rate of around 95%, 97%, and 99%, respectively, which is clearly higher than the column direction. Mapping the input data along the row not only achieves higher accuracy but also has less consumption of time as there are only 20 dimensions of data to be reduced. Same as the previous experimental setup, the overall accuracy rate in (10 × 20) is not as high as that of (20 × 60). However, in row direction experiments, in both cases ((10 × 20) and (20 × 60) sample sizes), the accuracy rate goes upper than that of column direction method with less time-consumption duration.
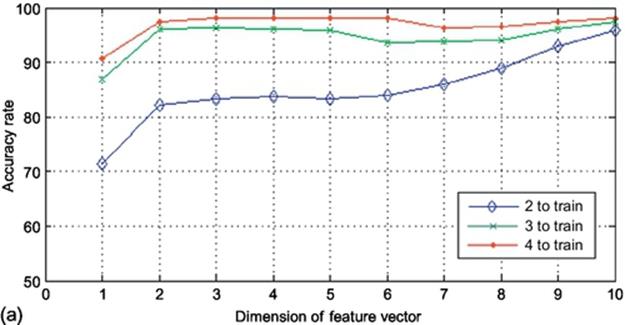
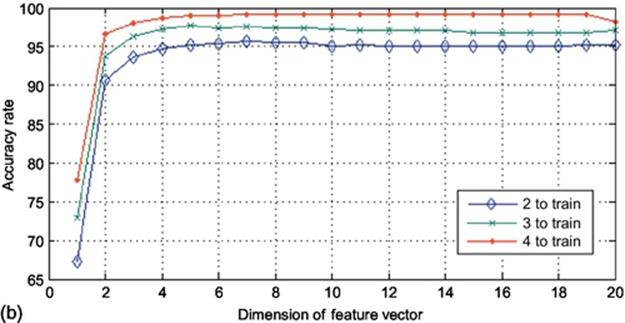
FIGURE 5 Accuracy rates obtained using K2DPCA in row direction on finger vein database: (a) (10 × 20) sample size and (b) (20 × 60) sample size.
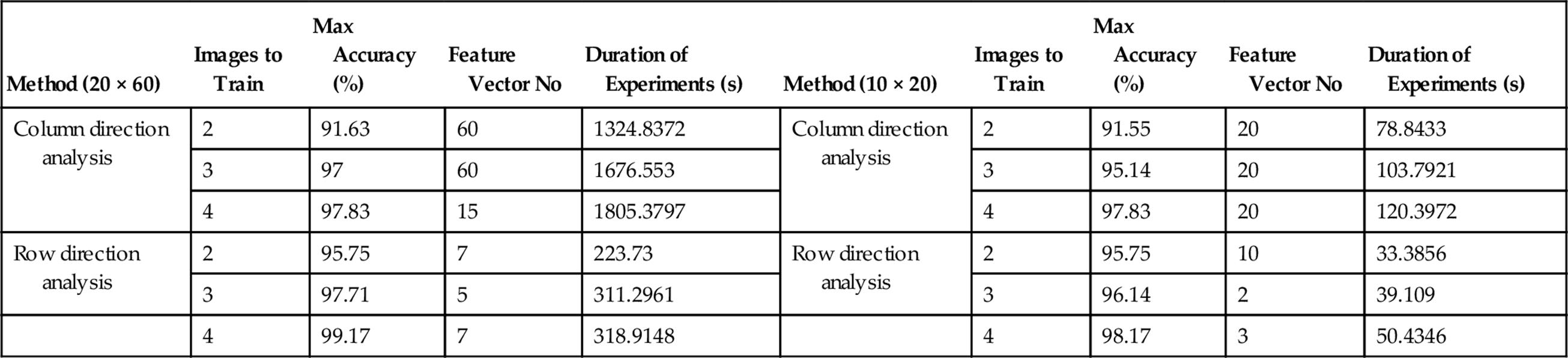
In this part, we give a summary of the whole experiments and their corresponding results for the sake of a better comparison. We have chosen the highest accuracies of each method in all implementations and their corresponding dimension of feature vector. All the mentioned information are indicated in Table 1 in addition to the duration of time each algorithm consumed to analyze the data. As Table 1 shows, the maximum accuracy of the row direction analysis is higher than that of column direction in all the different experiments. Furthermore, it is observed that not only it leads to higher accuracy, but also its dimension of feature vector is much less than that of column method in all implementations implying that the row direction method can be even faster than the column direction method in real-time system as it reaches the higher accuracy using less feature vectors.
Table 1
Comparison of the Proposed Algorithm in Row and Column Direction
7 Conclusion
In this paper, we have proposed a new method to enhance the performance of finger vein recognition and analyzed two different aspects of applying it in order to determine the most appropriate one. Our algorithm uses kernel mapping in two different directions to transfer the input data to another space where applying 2DPCA merits the final output of the system. We also used Euclidian distance as classifier in the last step of the algorithm. Extensive experiments were conducted on our database using three different numbers of images for training. Results demonstrate that mapping the data in row direction reaches both having higher accuracy and consuming less time compared to the column direction method meaning that the proposed method has the highest accuracy when mapping the data along the row direction.
References
[1] Jain AK, Ross A, Prabhakar S. An introduction to biometric recognition. IEEE T Circuit Syst. 2004;14(1):4–20.
[2] Schouten B, Jacobs B. Biometrics and their use in e-passports. Image Vis Comput. 2009;27(3):305–312.
[3] Beng TS, Rosdi BA. Finger-vein identification using pattern map and principal component analysis. In: 2011 IEEE international conference signal image processing applications, November; 2011:530–534.
[4] Kumar A, Zhou Y. Human identification using finger images. IEEE T Image Process. 2012;21(4):2228–2244.
[5] Zhao W, et al. Face recognition: A literature survey. Acm Computing Surveys (CSUR) 2003;35(4):399–458.
[6] Chellapa SR, Wilson CL, Sirohey S. Human and machine recognition of faces: a survey. Proc IEEE. 1995;83:705–740.
[7] Yang G, Xi X, Yin Y. Finger vein recognition based on a personalized best bit map. Sensors (Basel). 2012;12(2):1738–1757.
[8] Song W, Kim T, Kim HC, Choi JH, Kong H-J, Lee S-R. A finger-vein verification system using mean curvature. Pattern Recogn Lett. 2011;32(11):1541–1547.
[9] Yang J, Shi Y, Yang J. Personal identification based on finger-vein features. Comput Hum Behav. 2011;27(5):1565–1570.
[10] Wu J-D, Ye S-H. Driver identification using finger-vein patterns with radon transform and neural network. Expert Syst Appl. 2009;36(3):5793–5799.
[11] Wu J-D, Liu C-T. Finger-vein pattern identification using principal component analysis and the neural network technique. Expert Syst Appl. 2011;38(5):5423–5427.
[12] Bishop CM. Pattern recognition and machine learning. Vol. 1. New York: Springer, 2006.
[13] Kim KI, Jung K, Kim HJ. Face recognition using kernel principal component analysis. Signal Process. 2002;9(2):40–42.
[14] Ebied RM. Feature extraction using PCA and Kernel-PCA for face recognition. Int Conf Inform Syst. 2012;8:72–77.
[15] Hu P, Yang A. Indefinite Kernel entropy component analysis. Sci Tech. 2010;3:0–3.
[16] Jenssen R. Kernel entropy component analysis. IEEE Trans Pattern Anal Mach Intell. May 2010;32(5):847–860.
[17] Shekar BH, Sharmila Kumari M, Mestetskiy LM, Dyshkant NF. Face recognition using kernel entropy component analysis. Neurocomputing. 2011;74(6):1053–1057.
[18] Shawe-Taylor J, Cristianini N. Kernel methods for pattern analysis. New York, NY: Cambridge University Press; 2004 p. 462.
[19] Damavandinejadmonfared S, Mobarakeh AK, Suandi SA, Rosdi BA. Evaluate and determine the most appropriate method to identify finger vein. Procedia Eng. 2012;41:516–521.
[20] Damavandinejadmonfared S. Kernel entropy component analysis using local mean-based k-nearest centroid neighbour (LMKNCN) as a classifier for face recognition in video surveillance camera systems. In: IEEE international conference on intelligent computer communication and processing (ICCP); 2012:253–256.
[21] Damavandinejadmonfared S. Finger vein recognition using linear Kernel entropy component analysis. In: IEEE international conference on intelligent computer communication and processing (ICCP); 2012:249–252.
[22] Damavandinejadmonfared S, Varadharajan V. Effective Kernel mapping for one-dimensional principal component analysis in finger vein recognition. In: The 2014 international conference on image processing, computer vision, and pattern recognition; 2014.
[23] Hsieh P-C, Tung P-C. A novel hybrid approach based on sub-pattern technique and whitened PCA for face recognition. Pattern Recogn. 2009;42(5):978–984.
[24] Yang L. A human face recognition method by improved modular 2DPCA. In: IT in Medicine and Education (ITME), 2011 international symposium, vol. 2; 2011:7–11.
[25] Nhat VDM, Lee S. Kernel-based 2DPCA for face recognition. In: 2007 IEEE international symposium on signal processing and information technology, December; 2007:35–39.
[26] Yu C, Qing H, Zhang L. K2DPCA plus 2DPCA: an efficient approach for appearance based object recognition. In: 2009 3rd international conference on bioinformatics and biomedical engineering, June; 2009:1–4.
[27] Damavandinejadmonfared S, Varadharajan V. Finger vein recognition in row and column directions using two dimensional Kernel principal component analysis. In: The 2014 international conference on image processing, computer vision, and pattern recognition; 2014.